Hypericaceae) Heritiana S
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hypericaceae Key, Charts & Traits
Hypericaceae (St. Johnswort Family) Traits, Keys, & Comparison Charts © Susan J. Meades, Flora of Newfoundland and Labrador (Aug. 8, 2020) Hypericaceae Traits ........................................................................................................................ 1 Hypericaceae Key ........................................................................................................................... 2 Comparison Charts (3) ................................................................................................................... 4 References ...................................................................................................................................... 7 Hypericaceae Traits • Perennial herbs (in our area). • Stems are erect (lax in plants growing in flooded habitats) and glabrous; terete (round), or square in cross-section; internodes of terete stems with or without 2 low, vertical ridges along their length. • Leaves are cauline, opposite, and usually sessile; blades are simple, linear to ovate, with mostly entire margins; apices are obtuse to rounded; stipules are absent. • Pellucid glands with essential oils appear as translucent dots on the leaves (visible when leaves are held up to the light). • Dark red to blackish glands (with essential oils like hypericin) appear as slender streaks or tiny dots along the leaf, sepal, or petal margins of some species. • Flowers are solitary or 2–40 in terminal and often axillary simple to compound cymes, rarely in panicles. • Flowers are bisexual -

1. EUONYMUS Linnaeus, Sp. Pl. 1: 197
Fl. China 11: 440–463. 2008. 1. EUONYMUS Linnaeus, Sp. Pl. 1: 197. 1753 [“Evonymus”], nom. cons. 卫矛属 wei mao shu Ma Jinshuang (马金双); A. Michele Funston Shrubs, sometimes small trees, ascending or clambering, evergreen or deciduous, glabrous, rarely pubescent. Leaves opposite, rarely also alternate or whorled, entire, serrulate, or crenate, stipulate. Inflorescences axillary, occasionally terminal, cymose. Flowers bisexual, 4(or 5)-merous; petals light yellow to dark purple. Disk fleshy, annular, 4- or 5-lobed, intrastaminal or stamens on disk; anthers longitudinally or obliquely dehiscent, introrse. Ovary 4- or 5-locular; ovules erect to pendulous, 2(–12) per locule. Capsule globose, rugose, prickly, laterally winged or deeply lobed, occasionally only 1–3 lobes developing, loculicidally dehiscent. Seeds 1 to several, typically 2 developing, ellipsoid; aril basal to enveloping seed. Two subgenera and ca. 130 species: Asia, Australasia, Europe, Madagascar, North America; 90 species (50 endemic, one introduced) in China. Euonymus omeiensis W. P. Fang (J. Sichuan Univ., Nat. Sci. Ed. 1: 38. 1955) was described from Sichuan (Emei Shan, Shishungou, ca. 1300 m). This putative species was misdiagnosed; it is a synonym of Reevesia pubescens Masters in the Sterculiaceae (see Fl. China 12: 317. 2007). The protologue describes the fruit as having bracts. The placement of Euonymus tibeticus W. W. Smith (Rec. Bot. Surv. India 4: 264. 1911), described from Xizang (3000–3100 m) and also occurring in Bhutan (Lhakhang) and India (Sikkim), is unclear, as only a specimen with flower buds is available. Euonymus cinereus M. A. Lawson (in J. D. Hooker, Fl. Brit. India 1: 611. 1875) was described from India. -

Mario Gomes1
Rodriguésia 63(4): 1157-1163. 2012 http://rodriguesia.jbrj.gov.br Nota Científica / Short Communication Kielmeyera aureovinosa (Calophyllaceae) – a new species from the Atlantic Rainforest in highlands of Rio de Janeiro state Kielmeyera aureovinosa (Calophyllaceae) - uma nova espécie da Mata Atlântica na região serrana do estado do Rio de Janeiro Mario Gomes1 Abstract Kielmeyera aureovinosa M. Gomes is a tree of the Atlantic Rainforest, endemic to the highlands of Rio de Janeiro state, occurring in riverine forest. The new species is distinguished in the genus by having a wine colored stem with metallic luster, peeling, with golden bands: it differs from other species of Kielmeyera section Callodendron by having leaves with sparse resinous corpuscles and flowers with ciliate margined sepals and petals. This paper provides a description of the species, illustrations and digital images; morphological and palynological features of Kielmeyera section Callodendron species are discussed and compared. Key words: Calophyllaceae, Kielmeyera aureovinosa, Atlantic Rainforest, riverine forest, Rio de Janeiro state. Resumo Kielmeyera aureovinosa M.Gomes é uma árvore da Mata Atlântica, endêmica da região serrana do estado do Rio de Janeiro, ocorrente em matas ciliares. A nova espécie é distinta das demais no gênero por ter caule de coloração vinoso-metálica, desfolhante, com faixas e nuances dourados; diferencia-se das demais espécies de Kielmeyera seção Callodendron por possuir folhas com corpúsculos resiníferos esparsos e flores com sépalas e pétalas de margens ciliadas. Este trabalho fornece descrição da espécie, estado de conservação, ilustrações esquemáticas e imagens digitais; características morfológicas e palinológicas das espécies de Kielmeyera seção Callodendron são discutidas e apresentadas em tabelas para comparação. -

Facultad De Ciencias Forestales
FACULTAD DE CIENCIAS FORESTALES ESCUELA DE FORMACIÓN PROFESIONAL DE INGENIERÍA EN ECOLOGÍA DE BOSQUES TROPICALES TESIS “Cálculo del área foliar de Caraipa utilis Vásquez y su contribución para su manejo sostenible en los Varillales de la Reserva Nacional Allpahuayo Mishana, Loreto, Perú” Tesis para optar el título de Ingeniero en Ecología de Bosques Tropicales Autor Alan Christian Chumbe Ycomedes Iquitos - Perú 2017 DEDICATORIA A DIOS, por brindarme cada día un nuevo amanecer para ser una mejor persona. A mis padres, David y Janeth, y hermana, Giovanna, personas que son mi ejemplo a seguir y estímulo para ser mejor cada día. A mis amigos y demás personas que estuvieron presentes en el día a día, que con su ayuda y compañía me incentivaron a concluir este proyecto. AGRADECIMIENTO Al Blgo. Ricardo Zárate Gómez, por su paciencia y co-asesoramiento para lograr concluir el proyecto. Un agradecimiento especial al docente Fritz Veintemilla Arana, por su constante apoyo y asesoramiento, y los consejos brindados para el desarrollo de la presente tesis. A Luisin Ruiz, Max Guiriz, Priscila Gonzales, Danna Flores, Milagros Rimachi, Linder Mozombite y George Gallardo por su apoyo en el trabajo de campo. i ÍNDICE Pág. I. Introducción ............................................................................................... 1 II. El problema ................................................................................................ 3 2.1. Descripción del problema .................................................................. 3 2.2. Definición -

Celastraceae
Species information Abo ut Reso urces Hom e A B C D E F G H I J K L M N O P Q R S T U V W X Y Z Celastraceae Family Profile Celastraceae Family Description A family of about 94 genera and 1400 species, worldwide; 22 genera occur naturally in Australia. Genera Brassiantha - A genus of two species in New Guinea and Australia; one species occurs naturally in Australia. Simmons et al (2012). Celastrus - A genus of 30 or more species, pantropic; two species occur naturally in Australia. Jessup (1984). Denhamia - A genus of about 17 species in the Pacific and Australia; about 15 species occur in Australia. Cooper & Cooper (2004); McKenna et al (2011); Harden et al (2014); Jessup (1984); Simmons (2004). Dinghoua - A monotypic genus endemic to Australia. Simmons et al (2012). Elaeodendron - A genus of about 80 species, mainly in the tropics and subtropics particularly in Africa; two species occur naturally in Australia. Harden et al. (2014); Jessup (1984) under Cassine; Simmons (2004) Euonymus - A genus of about 180 species, pantropic, well developed in Asia; one species occur naturally in Australia. Hou (1975); Jessup (1984); Simmons et al (2012). Gymnosporia - A genus of about 100 species in the tropics and subtropics, particularly Africa; one species occurs naturally in Australia. Jordaan & Wyk (1999). Hedraianthera - A monotypic genus endemic to Australia. Jessup (1984); Simmons et al (2012). Hexaspora - A monotypic genus endemic to Australia. Jessup (1984). Hippocratea - A genus of about 100 species, pantropic extending into the subtropics; one species occurs naturally in Australia. -

Systematics and Floral Evolution in the Plant Genus Garcinia (Clusiaceae) Patrick Wayne Sweeney University of Missouri-St
University of Missouri, St. Louis IRL @ UMSL Dissertations UMSL Graduate Works 7-30-2008 Systematics and Floral Evolution in the Plant Genus Garcinia (Clusiaceae) Patrick Wayne Sweeney University of Missouri-St. Louis Follow this and additional works at: https://irl.umsl.edu/dissertation Part of the Biology Commons Recommended Citation Sweeney, Patrick Wayne, "Systematics and Floral Evolution in the Plant Genus Garcinia (Clusiaceae)" (2008). Dissertations. 539. https://irl.umsl.edu/dissertation/539 This Dissertation is brought to you for free and open access by the UMSL Graduate Works at IRL @ UMSL. It has been accepted for inclusion in Dissertations by an authorized administrator of IRL @ UMSL. For more information, please contact [email protected]. SYSTEMATICS AND FLORAL EVOLUTION IN THE PLANT GENUS GARCINIA (CLUSIACEAE) by PATRICK WAYNE SWEENEY M.S. Botany, University of Georgia, 1999 B.S. Biology, Georgia Southern University, 1994 A DISSERTATION Submitted to the Graduate School of the UNIVERSITY OF MISSOURI- ST. LOUIS In partial Fulfillment of the Requirements for the Degree DOCTOR OF PHILOSOPHY in BIOLOGY with an emphasis in Plant Systematics November, 2007 Advisory Committee Elizabeth A. Kellogg, Ph.D. Peter F. Stevens, Ph.D. P. Mick Richardson, Ph.D. Barbara A. Schaal, Ph.D. © Copyright 2007 by Patrick Wayne Sweeney All Rights Reserved Sweeney, Patrick, 2007, UMSL, p. 2 Dissertation Abstract The pantropical genus Garcinia (Clusiaceae), a group comprised of more than 250 species of dioecious trees and shrubs, is a common component of lowland tropical forests and is best known by the highly prized fruit of mangosteen (G. mangostana L.). The genus exhibits as extreme a diversity of floral form as is found anywhere in angiosperms and there are many unresolved taxonomic issues surrounding the genus. -

Reproductive Biology of <I>Pentadesma
Plant Ecology and Evolution 148 (2): 213–228, 2015 http://dx.doi.org/10.5091/plecevo.2015.998 REGULAR PAPER Reproductive biology of Pentadesma butyracea (Clusiaceae), source of a valuable non timber forest product in Benin Eben-Ezer B.K. Ewédjè1,2,*, Adam Ahanchédé3, Olivier J. Hardy1 & Alexandra C. Ley1,4 1Service Evolution Biologique et Ecologie, Faculté des Sciences, Université Libre de Bruxelles, 50 Av. F. Roosevelt, 1050 Brussels, Belgium 2Faculté des Sciences et Techniques FAST-Dassa, BP 14, Dassa-Zoumé, Université d’Abomey-Calavi, Benin 3Faculté des Sciences Agronomiques FSA, BP526, Université d’Abomey-Calavi, Benin 4Institut für Geobotanik und Botanischer Garten, University Halle-Wittenberg, Neuwerk 21, 06108 Halle (Saale), Germany *Author for correspondence: [email protected] Background and aims – The main reproductive traits of the native African food tree species, Pentadesma butyracea Sabine (Clusiaceae), which is threatened in Benin and Togo, were examined in Benin to gather basic data necessary to develop conservation strategies in these countries. Methodology – Data were collected on phenological pattern, floral morphology, pollinator assemblage, seed production and germination conditions on 77 adult individuals from three natural populations occurring in the Sudanian phytogeographical zone. Key results – In Benin, Pentadesma butyracea flowers once a year during the dry season from September to December. Flowering entry displayed less variation among populations than among individuals within populations. However, a high synchrony of different floral stages between trees due to a long flowering period (c. 2 months per tree), might still facilitate pollen exchange. Pollen-ovule ratio was 577 ± 213 suggesting facultative xenogamy. The apical position of inflorescences, the yellowish to white greenish flowers and the high quantity of pollen and nectar per flower (1042 ± 117 µL) represent floral attractants that predispose the species to animal-pollination. -

Vascular Plants of Pu'uhonua 0 Hiinaunau National
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by ScholarSpace at University of Hawai'i at Manoa Technical Report 105 Vascular Plants of Pu'uhonua 0 Hiinaunau National Historical Park Technical Report 106 Birds of Pu'uhonua 0 Hiinaunau National Historical Park COOPERATIVE NATIONAL PARK RESOURCES STUDIES UNIT UNIVERSITY OF HAWAI'I AT MANOA Department of Botany 3 190 Maile Way Honolulu, Hawai'i 96822 (808) 956-8218 Clifford W. Smith, Unit Director Technical Report 105 VASCULAR PLANTS OF PU'UHONUA 0 HONAUNAU NATIONAL HISTORICAL PARK Linda W. Pratt and Lyman L. Abbott National Biological Service Pacific Islands Science Center Hawaii National Park Field Station P. 0.Box 52 Hawaii National Park, HI 967 18 University of Hawai'i at Manoa National Park Service Cooperative Agreement CA8002-2-9004 May 1996 TABLE OF CONTENTS Page . LIST OF FIGURES ............................................. 11 ABSTRACT .................................................. 1 ACKNOWLEDGMENTS .........................................2 INTRODUCTION ..............................................2 THESTUDYAREA ............................................3 Climate ................................................ 3 Geology and Soils ......................................... 3 Vegetation ..............................................5 METHODS ...................................................5 RESULTS AND DISCUSSION .....................................7 Plant Species Composition ...................................7 Additions to the -

A New Species from Cerro De La Neblina, Venezuela Lucas C
ACTA AMAZONICA http://dx.doi.org/10.1590/1809-4392202000072 ORIGINAL ARTICLE Tovomita nebulosa (Clusiaceae), a new species from Cerro de la Neblina, Venezuela Lucas C. MARINHO1,2* , Manuel LUJÁN3, Pedro FIASCHI4, André M. AMORIM5,6 1 Universidade Federal do Maranhão, Centro de Ciências Biológicas e da Saúde, Departamento de Biologia, Av. dos Portugueses 1966, Bacanga 65080-805, São Luís, Maranhão, Brazil 2 Universidade Estadual de Feira de Santana, Programa de Pós-Graduação em Botânica, Av. Transnordestina s/n, Novo Horizonte 44036-900, Feira de Santana, Bahia, Brazil 3 California Academy of Sciences, 55 Music Concourse Drive, Golden Gate Park, San Francisco, California 94118, USA 4 Universidade Federal de Santa Catarina, Centro de Ciências Biológicas, Departamento de Botânica, Trindade 88040-900, Florianópolis, Santa Catarina, Brazil 5 Universidade Estadual de Santa Cruz, Departamento de Ciências Biológicas, Km 25 Rodovia Ilhéus-Itabuna 45662-900, Ilhéus, Bahia, Brazil 6 Centro de Pesquisas do Cacau, Herbário CEPEC, Km 16 Rodovia Itabuna-Ilhéus 45650-970, Itabuna, Bahia, Brazil * Corresponding author: [email protected]; https://orcid.org/0000-0003-1263-3414 ABSTRACT Although the number of recently described Tovomita species is relatively high, much more remains to be done, given that each new survey of representative Amazonian collections reveals many potentially undescribed taxa. In the treatment for Tovomita published in Flora of the Venezuelan Guayana, at least six distinct morphotypes did not match any previously described species. -

Hypericum Gentianoides (L.) BSP Gentian-Leaved St. John's-Wort
Hypericum gentianoides (L.) B.S.P. gentian-leavedgentian-leaved St. John’s-wortSt. John’s-wort, Page 1 State Distribution Photo by Susan R. Crispin Best Survey Period Jan Feb Mar Apr May Jun Jul Aug Sept Oct Nov Dec Status: State special concern of the occurrences distributed in Wayne, Monroe, Van Buren, and St. Clair counties. Global and state rank: G5/S3 Recognition: H. gentianoides is an annual species Other common names: orange-grass, orange-grass St. ranging from 1-2 dm in height or more, with an erect, John’s-wort central stem that terminates in a number of slender, strongly ascending branches. When crushed, the plant Family: Clusiaceae (St. John’s-wort family); also produces a faint, citrus-like fragrance (which has known as the Guttiferae, and sometimes placed in the also been described as a peach-like odor), as indicated separate family Hypericaceae, similarly long known as by some of the common names for this species. The the St. John’s-wort family. tiny, linear leaves, which are opposite and appressed (oriented to be parallel with the stem), are highly Synonym: Sarothra gentianoides L. reduced, scale-like, and less than 3 mm long. The yellow, five-parted flowers, which areusually solitary Taxonomy: Long treated as a separate family, the in the upper leaf axils, are less than 3 mm broad, Hypericaceae is now combined with the Clusiaceae have 3 styles, and bear fewer than 100 stamens. The (Guttiferae) by most recent treatments “stick-like” appearance of this St. John’s-wort, including the minute, linear leaves, makes it unlikely that this Range: Primarily occurring in eastern North America, will be confused with another species. -
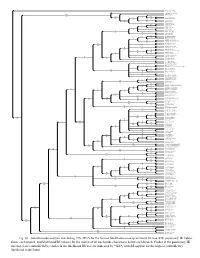
Fig. S1. Simultaneous-Analysis (Excluding ITS Rdna for the Former
Lepidobotrys staudtii Ruptiliocarpon caracolito Lepuropetalon spathulatum 100 Parnassia faberi 100 95 Parnassia glauca 100 100 Parnassia palustris Fay 78 86 100 100 Parnassia palustris 2600 81 98 100 Parnassia tenella 97 100 Parnassia trinervis Parnassia laxmannii 52 Allocassine laurifolia 77 *50* Lauridia reticulata 84 Lauridia tetragona 97 50 Cassine peragua 58 98 Cassine schinoides Maurocenia frangula 97 100 Catha edulis 1896 100 Catha edulis 3016 89 Gymnosporia arbutifolia 51 90 Gymnosporia royleana 58 Gymnosporia engleriana 100 Gymnosporia cassinoides 2201 100 Gymnosporia cassinoides 3525 99 100 Gymnosporia diversifolia 99 Gymnosporia inermis 100 100 Gymnosporia harveyana 99 100 Gymnosporia mossambicensis 98 100 Gymnosporia linearis 100 100 Gymnosporia senegalensis 3030 99 97 Gymnosporia pyria 88 95 Gymnosporia senegalensis 4 93 50 Gymnosporia polyacantha 89 96 98 Putterlickia pyracantha Putterlickia verrucosa 73 99 71 Gymnosporia divaricata 71 Lydenburgia abbottii 52 Lydenburgia cassinoides Empleuridium juniperinum 56 Maytenus cordata Mystroxylon aethiopicum subsp macrocarpum 57 72 80 Maytenus procumbens 100 100 82 Maytenus undata 64700 100 100 Maytenus undata 3028 Pseudosalacia streyi 100 Pterocelastrus echinatus 99 Pterocelastrus tricuspidatus Robsonodendron eucleiforme 91 Brexia madagascariensis 92 89 Polycardia phyllanthoides 100 86 Polycardia libera 100 Polycardia lateralis 93 Elaeodendron australe var angustifolia 100 91 Elaeodendron australe var integrifolium 100 100 Elaeodendron melanocarpum 57 100 Elaeodendron cunninghamii -

Calophyllaceae
DOI: 10.13102/scb884 ARTIGO Flora da Bahia: Calophyllaceae Amanda Pricilla Batista Santos*, Fabio da Silva do Espírito Santoa & Alessandro Rapinib Programa de Pós-graduação em Botânica, Departamento de Ciências Biológicas, Universidade Estadual de Feira de Santana, Feira de Santana, Bahia, Brasil. Resumo – É apresentado aqui o tratamento taxonômico de Calophyllaceae para o estado da Bahia, Brasil. São reconhecidos quatro gêneros e 21 espécies: Calophyllum (C. brasiliense), Caraipa (C. densifolia), Kielmeyera (18 espécies) e Mammea (M. americana), das quais sete espécies são endêmicas do estado. São apresentadas chaves de identificação para os gêneros e espécies, descrições, comentários taxonômicos, ilustrações e mapas de distribuição geográfica das espécies no estado. Palavras-chave adicionais: florística, Kielmeyera, Nordeste do Brasil, taxonomia. Abstract (Flora of Bahia: Calophyllaceae) – The taxonomic treatment of the Calophyllaceae from Bahia state, Brazil, is presented here. Four genera and 21 species are recognized: Calophyllum (C. brasiliense), Caraipa (C. densifolia), Kielmeyera (18 species) and Mammea (M. americana), of which seven species are endemic to the state. Identification keys to genera and species, descriptions, taxonomic comments, illustrations and maps of geographic distribution of species in the state are presented. Additional keywords: floristics, Kielmeyera, Northeast Brazil, taxonomy. CALOPHYLLACEAE clusioides (Malpighiales), Calophyllaceae está mais relacionada com o clado composto por Podostemaceae Árvores, arbustos, raramente lianas, latescentes, e Hypericaceae, enquanto Clusiaceae s.s. parece estar monoicas, poligâmicas ou dioicas. Folhas alternas ou mais relacionada com Bonnetiaceae (Xi et al. 2012). opostas, simples, sem estípulas, margem inteira. Flores mono ou diclinas, actinomorfas, diclamídeas ou Chave para gêneros raramente monoclamídeas (412 tépalas), prefloração 1. Folhas alternas; frutos capsulares.