Allozymic Variation in Four Indian Species of Genus Mus:A Comparative Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evolutionary Characterization of a Y Chromosomal Sequence Conserved in the Genus Mus
Genet. Res., Camb. (1988), 52, pp. 145-150 With 5 text-figures Printed in Great Britain 145 Evolutionary characterization of a Y chromosomal sequence conserved in the genus Mus YUTAKA NISHIOKA Department of Biology, McGill University, Montreal, Quebec, Canada H3A IBI (Received 8 October 1987 and in revised form 26 January 1988) Summary The extent of accumulation of mouse Y chromosomal repetitive sequences generally correlates with the known phylogenetic relationships in the genus Mus. However, we describe here a M. musculus Y chromosomal repetitive sequence, designated as ACClfl, whose accumulation patterns among eight Mus species do not correspond to their phylogenetic relationships. Although male-specific hybridization bands were present in all the species examined, significant accumulation (^ 200 copies) in the Y chromosomes was found in M. minutoides (subgenus Nannomys), M. pahari (subgenus Coelomys) and M. saxicola (subgenus Pyromys) as well as in the three closely related species M. hortulanus, M. musculus and M. spretus that belong to the subgenus Mus. Unexpectedly, the Y chromosomes of M. caroli and M. cookii (both subgenus Mus) had considerably reduced amounts of ACClfl-related sequences. Furthermore, in rats {Rattus norvegicus) the major accumulation sites appear to be autosomal. These observations suggest that caution must be taken in the interpretation of data obtained with repetitive sequences that have evolved quickly. 1. Introduction over 50 mouse (M. musculus) Y chromosomal sequences (Nishioka & Lamothe, 1987a). To date, 32 Recently several groups have isolated mouse DNA fragments were generated from 11 original (Mus musculus) Y chromosomal repetitive sequences isolates and their conservation in the genus Mus has (Bishop et al. -

Genus/Species Skull Ht Lt Wt Stage Range Abalosia U.Pliocene S America Abelmoschomys U.Miocene E USA A
Genus/Species Skull Ht Lt Wt Stage Range Abalosia U.Pliocene S America Abelmoschomys U.Miocene E USA A. simpsoni U.Miocene Florida(US) Abra see Ochotona Abrana see Ochotona Abrocoma U.Miocene-Recent Peru A. oblativa 60 cm? U.Holocene Peru Abromys see Perognathus Abrosomys L.Eocene Asia Abrothrix U.Pleistocene-Recent Argentina A. illuteus living Mouse Lujanian-Recent Tucuman(ARG) Abudhabia U.Miocene Asia Acanthion see Hystrix A. brachyura see Hystrix brachyura Acanthomys see Acomys or Tokudaia or Rattus Acarechimys L-M.Miocene Argentina A. minutissimus Miocene Argentina Acaremys U.Oligocene-L.Miocene Argentina A. cf. Murinus Colhuehuapian Chubut(ARG) A. karaikensis Miocene? Argentina A. messor Miocene? Argentina A. minutissimus see Acarechimys minutissimus Argentina A. minutus Miocene? Argentina A. murinus Miocene? Argentina A. sp. L.Miocene Argentina A. tricarinatus Miocene? Argentina Acodon see Akodon A. angustidens see Akodon angustidens Pleistocene Brazil A. clivigenis see Akodon clivigenis Pleistocene Brazil A. internus see Akodon internus Pleistocene Argentina Acomys L.Pliocene-Recent Africa,Europe,W Asia,Crete A. cahirinus living Spiny Mouse U.Pleistocene-Recent Israel A. gaudryi U.Miocene? Greece Aconaemys see Pithanotomys A. fuscus Pliocene-Recent Argentina A. f. fossilis see Aconaemys fuscus Pliocene Argentina Acondemys see Pithanotomys Acritoparamys U.Paleocene-M.Eocene W USA,Asia A. atavus see Paramys atavus A. atwateri Wasatchian W USA A. cf. Francesi Clarkforkian Wyoming(US) A. francesi(francesci) Wasatchian-Bridgerian Wyoming(US) A. wyomingensis Bridgerian Wyoming(US) Acrorhizomys see Clethrionomys Actenomys L.Pliocene-L.Pleistocene Argentina A. maximus Pliocene Argentina Adelomyarion U.Oligocene France A. vireti U.Oligocene France Adelomys U.Eocene France A. -

Evolutionary History of the Subgenus Mus in Eurasia with Special Emphasis on the House Mouse Mus Musculus
Papers in Honour of Ken Aplin edited by Julien Louys, Sue O’Connor and Kristofer M. Helgen Helgen, Kristofer M., Julien Louys, and Sue O’Connor. 2020. The lives of creatures obscure, misunderstood, and wonderful: a volume in honour of Ken Aplin 1958–2019 ..........................149 Armstrong, Kyle N., Ken Aplin, and Masaharu Motokawa. 2020. A new species of extinct False Vampire Bat (Megadermatidae: Macroderma) from the Kimberley Region of Western Australia ........................................................................................................... 161 Cramb, Jonathan, Scott A. Hocknull, and Gilbert J. Price. 2020. Fossil Uromys (Rodentia: Murinae) from central Queensland, with a description of a new Middle Pleistocene species ............................................................................................................. 175 Price, Gilbert J., Jonathan Cramb, Julien Louys, Kenny J. Travouillon, Eleanor M. A. Pease, Yue-xing Feng, Jian-xin Zhao, and Douglas Irvin. 2020. Late Quaternary fossil vertebrates of the Broken River karst area, northern Queensland, Australia ........................ 193 Theden-Ringl, Fenja, Geoffrey S. Hope, Kathleen P. Hislop, and Benedict J. Keaney. 2020. Characterizing environmental change and species’ histories from stratified faunal records in southeastern Australia: a regional review and a case study for the early to middle Holocene ........................................................................................... 207 Brockwell, Sally, and Ken Aplin. 2020. Fauna on -

Evolutionary Characterization of a Y Chromosomal Sequence Conserved in the Genus Mus
Genet. Res., Camb. (1988), 52, pp. 145-150 With 5 text-figures Printed in Great Britain 145 Evolutionary characterization of a Y chromosomal sequence conserved in the genus Mus YUTAKA NISHIOKA Department of Biology, McGill University, Montreal, Quebec, Canada H3A IBI (Received 8 October 1987 and in revised form 26 January 1988) Summary The extent of accumulation of mouse Y chromosomal repetitive sequences generally correlates with the known phylogenetic relationships in the genus Mus. However, we describe here a M. musculus Y chromosomal repetitive sequence, designated as ACClfl, whose accumulation patterns among eight Mus species do not correspond to their phylogenetic relationships. Although male-specific hybridization bands were present in all the species examined, significant accumulation (^ 200 copies) in the Y chromosomes was found in M. minutoides (subgenus Nannomys), M. pahari (subgenus Coelomys) and M. saxicola (subgenus Pyromys) as well as in the three closely related species M. hortulanus, M. musculus and M. spretus that belong to the subgenus Mus. Unexpectedly, the Y chromosomes of M. caroli and M. cookii (both subgenus Mus) had considerably reduced amounts of ACClfl-related sequences. Furthermore, in rats {Rattus norvegicus) the major accumulation sites appear to be autosomal. These observations suggest that caution must be taken in the interpretation of data obtained with repetitive sequences that have evolved quickly. 1. Introduction over 50 mouse (M. musculus) Y chromosomal sequences (Nishioka & Lamothe, 1987a). To date, 32 Recently several groups have isolated mouse DNA fragments were generated from 11 original (Mus musculus) Y chromosomal repetitive sequences isolates and their conservation in the genus Mus has (Bishop et al. -

Download Article (PDF)
OCCASION P PER No. 297 Records of the Zoological Survey of ndia Li t of valid Rodent taxa (Class: Ma malia, Order: Rodentia) from Indian Subcontinent includ· g Myanmar M.S. PRAD AN AND S.S. TALMALE ZOOLOGIC L SURVEY OF I ' DIA OCCASIONAL PAPER No. 297 RECORDS OF THE ZOOLOGICAL SURVEY OF INDIA List of valid Rodent taxa (Class: Mammalia, Order: Rodentia) from Indian Subcontinent including Myanmar M.S. PRADHANI AND S.S. TALMALE2 Zoological Survey of India Western Regional Centre, Vidyanagar, Sector 29, Rawet Road PCNTDA Post, Pune, Maharashtra 411 044 Email: [email protected][email protected] Edited by the Director, Zoological Survey of India, Kolkata ~m Zoological Survey of India Kolkata CITATION Pradhan, M.S. and Talmale, S.S. 2009. List of valid Rodent taxa (Class : Mammalia; Order : Rodentia) from Indian Subcontinent including Myanmar, Rec. zool. Surv. India, Gcc. Paper No. 297 : 1-239. (Published by the Director, Zool. Surv. India, Kolkata) Published : October, 2009 ISBN J78-81-8171-224-0 t; Gnv!. of India, 2009 ALL RIGHTS RESERVED • No Part of this publication may be reproduced, stored in a retrieval system or transmitted in any form or by any means, electronic, mechanical, photocopying, recording or otherwise without the prior permission of the publisher. • This book is sold subject to the condition that it shall not, by way of trade, be lent, resold, hired out or otherwise disposed off without the publisher's consent, in a form of binding or cover other than that in which, it is published. • The correct price of this publication is the price printed on this page. -

The Testis-Determining Gene, SRY, Exists in Multiple Copies in Old World Rodents
Genet. Res., Camb. (1994), 64, pp. 151-159 With 6 text-figures Copyright © 1994 Printed in Great Britain 151 The testis-determining gene, SRY, exists in multiple copies in Old World rodents CLAUDE M. NAGAMINE Department of Cell Biology, Vanderbilt University School of Medicine, Medical Center North, Room T2216, Nashville, TN USA 37232-2175 (Received 19 July 1994) Summary SR Y is a unique gene on the Y chromosome in most mammalian species including the laboratory mouse, Mus musculus, and the closely related European wild mouse species M. spicilegus, M. macedonicus, and M. spretus. In contrast, SR Y is present in 2-6 copies in the more distantly related Asian mouse species M. caroli, M. cervicolor, and M. cookii and in 2—13 copies in the related murid species Pyromys saxicola, Coelomys pahari, Nannomys minutoides, Mastomys natalensis, and Rattus norvegicus. Copy numbers do not correlate with known phylogenetic relationships suggesting that SR Y has undergone a rapid and complex evolution in these species. SRY was recently proposed as a molecular probe for phylogenetic inferences. The presence of multiple SR Y genes in a wide range of murid species and genera, and at least one cricetid species, necessitates caution in the use of SR Y for phylogenetic studies in the Rodentia unless it is ascertained that multiple SR Y genes do not exist. family whose signature or characteristic amino acid 1. Introduction pattern is a DNA-binding domain of approximately Mammalian sex determination pivots on the absence 85 amino acids designated the HMG domain. The or presence of a Y chromosome. -
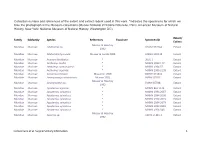
Collection Number and References of the Extant and Extinct Rodent Used in This Work
Collection number and references of the extant and extinct rodent used in this work. *indicates the specimens for which we take the photograph in the Museum collections (Musée National d’Histoire Naturelle, Paris; American Museum of Natural History, New York; National Museum of Natural History, Washington DC). Extant/ Family Subfamily Species References Fossil site Specimen ID Extinct Musser & Headley Muridae Murinae Abditomys sp. USNM 357244 Extant 1992 Muridae Murinae Abelomelomys sevia Musser & Lunde 2009 AMNH 192119 Extant Muridae Murinae Acomys dimidiatus * 2001 3 Extant Muridae Murinae Aethomys hindei * MNHN 1999 177 Extant Muridae Murinae Aethomys namanquesis * MNHN 1964 57 Extant Muridae Murinae Aethomys nigeriae * MNHN 1996 2239 Extant Muridae Murinae Anisomys imitator Missonne 1969 BMNH 471310 Extant Muridae Murinae Anonymomys mindorensis Musser 1981 FMNH 87597 Extant Musser & Headley Muridae Murinae Anonymomys sp. FMNH 87598 Extant 1992 Muridae Murinae Apodemus agrarius * MNHN BL6 1134 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2667 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2668 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2671 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2679 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2681 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 945 Extant Musser & Headley Muridae Murinae Apomys sp. 12971 CLM1-3 Extant 1992 Gomez Cano et al. Supplementary Information 1 Collection number and references of the extant and extinct rodent used in this work. *indicates the specimens for which we take the photograph in the Museum collections (Musée National d’Histoire Naturelle, Paris; American Museum of Natural History, New York; National Museum of Natural History, Washington DC). Extant/ Family Subfamily Species References Fossil site Specimen ID Extinct Musser & Headley Muridae Murinae Archboldomys sp. -

1 the House Mouse and Its Relatives
Cambridge University Press 978-0-521-76066-9 - Evolution of the House Mouse Edited by Miloš Macholán, Stuart J. E. Baird, Pavel Munclinger and Jaroslav Piálek Excerpt More information 1 The house mouse and its relatives: systematics and taxonomy JEAN-CHRISTOPHE AUFFRAY AND JANICE BRITTON-DAVIDIAN Introduction A quarter of a century before the advent of biochemical tools for taxo- nomic approaches, Schwarz and Schwarz (1943) published a pioneering survey of the systematics of the house mouse. All taxa were then considered as subspecies of the house mouse (Mus musculus). In other words, there was only one species within the subgenus Mus. It was a relatively large-scale study, but the taxonomic criteria were obviously based solely on external morphology and distribution of species. It is nowadays well established that the subgenus Mus exhibits high inter- specific morphological conservatism and that all species share an important part of their intra-specific variation. Most, if not all, classical external criteria such as coat colour, length of head, body, tail, ear, or foot, are poorly discriminating between species, and, except for very few (e.g. the zygomatic index for M. musculus;Orsini et al., 1983), none has a general and specific diagnostic value. When only one or two species occur sympatrically, some characters may be diagnostic, such as the length of the tail, which is useful to distinguish Mus musculus domesticus, one of the longest-tailed mice, from all sympatric species. Thus, there is only a poor corre- spondence between the 15 subspecies of Mus musculus described by Schwarz and Schwarz (1943) and the species that are now diagnosed by taxonomists. -

Influence of Predator and Food Chemical Cues in the Behaviour of the House Mouse (Mus Musculus)
En vue de l'obtention du DOCTORAT DE L'UNIVERSITÉ DE TOULOUSE Délivré par : Institut National Polytechnique de Toulouse (Toulouse INP) Discipline ou spécialité : Pathologie, Toxicologie, Génétique et Nutrition Présentée et soutenue par : M. CARLOS GRAU PARICIO le vendredi 11 janvier 2019 Titre : Influence of predator and food chemical cues in the behaviour of the house mouse (Mus musculus) Ecole doctorale : Sciences Ecologiques, Vétérinaires, Agronomiques et Bioingénieries (SEVAB) Unité de recherche : Département Sciences Agronomiques et Agroalimentaires (SSA-EIP) Directeur(s) de Thèse : M. PATRICK PAGEAT M. XAVIER MANTECA Rapporteurs : M. ANGELO GAZZANO, UNIVERSITA DI PISA Mme CHRISTINA BUESCHING, UNIVERSITY OF OXFORD Membre(s) du jury : M. DAVID MORTON, UNIVERSITY OF BIRMINGHAM, Président Mme CECILE BIENBOIRE-FROSINI, IRSEA, Membre M. PATRICK PAGEAT, IRSEA, Membre ACKNOWLEDGEMENTS/ REMERCIMENTS Je voudrais faire ces remerciements en français car c’est la langue maternelle de la grande majorité des personnes avec lesquelles j’ai travaillé pendant ces années de thèse et cette belle région du Luberon dans laquelle se trouve l’IRSEA. D’abord je voudrais remercier spécialement mon directeur de thèse Patrick Pageat, qui m’a guidé dans un premier temps comme chef de département et après comme directeur de thèse. Ce chemin que nous avons parcouru ensemble m’a permis acquérir l’expérience et les bons outils pour devenir chercheur et mûrir comme personne. Alessandro Cozzi m’a aidé d’abord comme chef de département et ensuite comme directeur de l’IRSEA. Julien Leclercq est la personne avec laquelle j’ai passé le plus de temps autour du travail de cette thèse. J’ai eu la chance de découvrir ses qualités au travail mes également ses qualités personnelles. -

Sri Lanka, 2019
Sri Lanka 7th – 20th February 2019 Trip report by John Wright. Email [email protected] Team: Nigel Goodgame, Sissel Goodgame, Nick Roberts, John Wright. Background We used the very well-known Bird and Wildlife Team who lived up to their amazing reputation for providing a high quality service with knowledgeable and dedicated naturalists. Our guide Dulan Vidanapathirana was no exception - he was brilliant, amazing in fact, his spotlighting ability was uncanny, he astounded us on more than one occasion and without doubt found most of the nocturnal animals. As well as mammals he is also an expert when it comes to the identification of birds, reptiles and amphibians – in fact he had a paper published on a new species of snake he discovered in Sri Lanka. Importantly, he is also great company, we all thoroughly recommend Dulan and we wouldn’t hesitate to use him again. Our driver, Herath, was good company, a careful driver and more than willing to do ‘odd’ hours us mammal watchers are apt to keep! He drove a comfortable 8-seater van which afforded us plenty of room. Bird and Wildlife Team see: https://www.birdandwildlifeteam.com/ We flew overnight direct from London Heathrow arriving in Colombo on Thursday 7th February. We flew back to London Heathrow on Wednesday 20th February on a direct flight from Colombo. On this trip we concentrated on seeing the many nocturnal mammals of Sri Lanka with Fishing Cat and Rusty-spotted Cat being top of the list of most wanted. Being all round naturalists (to some extent!) we of course didn’t ignore other forms of wildlife and recorded the birds, reptiles and amphibians we saw but struggled a bit with butterflies and dragonflies. -
Molecular Pattern Recognition of A
Hyperconservation of the N-Formyl Peptide Binding Site of M3: Evidence that M3 Is an Old Eutherian Molecule with Conserved Recognition of a Pathogen-Associated This information is current as Molecular Pattern of September 24, 2021. C. Kuyler Doyle, Beckley K. Davis, Richard G. Cook, Robert R. Rich and John R. Rodgers J Immunol 2003; 171:836-844; ; doi: 10.4049/jimmunol.171.2.836 Downloaded from http://www.jimmunol.org/content/171/2/836 References This article cites 63 articles, 25 of which you can access for free at: http://www.jimmunol.org/ http://www.jimmunol.org/content/171/2/836.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 24, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2003 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Hyperconservation of the N-Formyl Peptide Binding Site of M3: Evidence that M3 Is an Old Eutherian Molecule with Conserved Recognition of a Pathogen-Associated Molecular Pattern1 C. -

An Ancient Mammalian Gene Evidence That Thymus Leukemia
Hyperconservation of the Putative Antigen Recognition Site of the MHC Class I-b Molecule TL in the Subfamily Murinae: Evidence That Thymus Leukemia Antigen Is This information is current as an Ancient Mammalian Gene of September 24, 2021. Beckley K. Davis, Richard G. Cook, Robert R. Rich and John R. Rodgers J Immunol 2002; 169:6890-6899; ; doi: 10.4049/jimmunol.169.12.6890 Downloaded from http://www.jimmunol.org/content/169/12/6890 References This article cites 57 articles, 20 of which you can access for free at: http://www.jimmunol.org/ http://www.jimmunol.org/content/169/12/6890.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 24, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2002 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Hyperconservation of the Putative Antigen Recognition Site of the MHC Class I-b Molecule TL in the Subfamily Murinae: Evidence That Thymus Leukemia Antigen Is an Ancient Mammalian Gene1 Beckley K.