Characterization of Leptin Signaling in the Developing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Zebrafish Disease Models to Study the Pathogenesis of Inherited Manganese Transporter Defects and Provide A
Zebrafish disease models to study the pathogenesis of inherited manganese transporter defects and provide a route for drug discovery Dr Karin Tuschl University College London PhD Supervisors: Dr Philippa Mills & Prof Stephen Wilson A thesis submitted for the degree of Doctor of Philosophy University College London August 2016 Declaration I, Karin Tuschl, confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. Part of the work of this thesis has been published in the following articles for which copyright clearance has been obtained (see Appendix): - Tuschl K, et al. Manganese and the brain. Int Rev Neurobiol. 2013. 110:277- 312. - Tuschl K, et al. Mutations in SLC39A14 disrupt manganese homeostasis and cause childhood-onset parkinsonism-dystonia. Nat Comms. 2016. 7:11601. I confirm that these publications were written by me and may therefore partly overlap with my thesis. 2 Abstract Although manganese is required as an essential trace element excessive amounts are neurotoxic and lead to manganism, an extrapyramidal movement disorder associated with deposition of manganese in the basal ganglia. Recently, we have identified the first inborn error of manganese metabolism caused by mutations in SLC30A10, encoding a manganese transporter facilitating biliary manganese excretion. Treatment is limited to chelation therapy with intravenous disodium calcium edetate which is burdensome due to its route of administration and associated with high socioeconomic costs. Whole exome sequencing in patients with inherited hypermanganesaemia and early- onset parkinsonism-dystonia but absent SLC30A10 mutations identified SLC39A14 as a novel disease gene associated with manganese dyshomeostasis. -

Analyses of Allele-Specific Gene Expression in Highly Divergent
ARTICLES Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance James J Crowley1,10, Vasyl Zhabotynsky1,10, Wei Sun1,2,10, Shunping Huang3, Isa Kemal Pakatci3, Yunjung Kim1, Jeremy R Wang3, Andrew P Morgan1,4,5, John D Calaway1,4,5, David L Aylor1,9, Zaining Yun1, Timothy A Bell1,4,5, Ryan J Buus1,4,5, Mark E Calaway1,4,5, John P Didion1,4,5, Terry J Gooch1,4,5, Stephanie D Hansen1,4,5, Nashiya N Robinson1,4,5, Ginger D Shaw1,4,5, Jason S Spence1, Corey R Quackenbush1, Cordelia J Barrick1, Randal J Nonneman1, Kyungsu Kim2, James Xenakis2, Yuying Xie1, William Valdar1,4, Alan B Lenarcic1, Wei Wang3,9, Catherine E Welsh3, Chen-Ping Fu3, Zhaojun Zhang3, James Holt3, Zhishan Guo3, David W Threadgill6, Lisa M Tarantino7, Darla R Miller1,4,5, Fei Zou2,11, Leonard McMillan3,11, Patrick F Sullivan1,5,7,8,11 & Fernando Pardo-Manuel de Villena1,4,5,11 Complex human traits are influenced by variation in regulatory DNA through mechanisms that are not fully understood. Because regulatory elements are conserved between humans and mice, a thorough annotation of cis regulatory variants in mice could aid in further characterizing these mechanisms. Here we provide a detailed portrait of mouse gene expression across multiple tissues in a three-way diallel. Greater than 80% of mouse genes have cis regulatory variation. Effects from these variants influence complex traits and usually extend to the human ortholog. Further, we estimate that at least one in every thousand SNPs creates a cis regulatory effect. -

Peripheral Nervous System……………………………………………….9
PhD School in Integrative Biomedical Research Department of Pharmacological and Biomolecular Sciences Curriculum: Neuroscience Molecular basis for the development of innovative therapies for peripheral neuropathies treatment: role and cross-regulation of the GABAergic system and neuroactive steroids SDD BIO/09 - Physiology Luca Franco Castelnovo Badge number R10402 PhD Tutor: Prof. Valerio Magnaghi PhD School Coordinator: Prof. Chiarella Sforza Academic year 2015/2016 INDEX Abstract…………………………………………………………………...page 1 Abbreviations list…………………………………………………………….....5 Introduction………………………………………………………………….....8 Peripheral nervous system……………………………………………….9 General concepts……………...……….……………………………........……..9 Sensory system and nociceptive fibers.………………..………….…………..12 Schwann cells and myelination……….……..………………...………………15 Peripheral neuropathies…….…………………………………………...26 General concepts……………………………………………………...……….26 Neuropathic pain……………………………………...……………………….27 Nerve regeneration…………………………………………………………….28 The GABAergic system………….……………………………………...32 GABA……………….…………………………………………………..……..32 GABA-A receptors…………………………………………………………….33 GABA-B receptors…………………………………………………………….42 GABAergic system in the peripheral nervous system…………………………47 Protein kinase C – type ε……………………….………………………..51 General concepts…………………………………………………….…………51 Cross-talk with allopregnanolone and GABA-A………………………………54 Neuroactive steroids…………......………………………………………57 General concepts…………………………………….…………………………57 Mechanism of action…………………………….……………………………..59 Progesterone derivatives………………………………….……………………60 -

HPSE2 Gene Heparanase 2 (Inactive)
HPSE2 gene heparanase 2 (inactive) Normal Function The HPSE2 gene provides instructions for making a protein called heparanase 2. Little is known about this protein, but its structure is similar to that of another protein called heparanase 1. Heparanase 1 is an enzyme that splits (cleaves) molecules called heparan sulfate proteoglycans (HSPGs) by removing the heparan sulfate portion (the side chain). HSPGs are important parts of the lattice of proteins and other molecules outside the cell (extracellular matrix) and of basement membranes, which are thin, sheet-like structures that separate and support cells in many tissues. Cleavage of HSPGs by heparanase 1 may lead to changes in the basement membrane or extracellular matrix that allow cell movement or release of substances from the cell. The specific function of the heparanase 2 enzyme is not well understood, but studies suggest that it may block the action of heparanase 1. Health Conditions Related to Genetic Changes Migraine MedlinePlus Genetics provides information about Migraine Ochoa syndrome At least nine HPSE2 gene mutations have been identified in people with Ochoa syndrome (also called urofacial syndrome), a disorder that causes urinary problems and unusual facial expressions. These mutations result in changes in the heparanase 2 protein that likely prevent it from functioning. The connection between HPSE2 gene mutations and the features of Ochoa syndrome are unclear. Because the areas of the brain that control facial expression and urination are in close proximity, some researchers have suggested that the genetic changes may lead to an abnormality in this brain region that may account for the symptoms of Ochoa syndrome. -

Pathways and Networks Biological Meaning of the Gene Sets
Pathways and Networks Biological meaning of the gene sets Gene ontology terms ? Pathway mapping Linking to Pubmed abstracts or associted MESH terms Regulation by the same transcription factor (module) Protein families and domains Gene set enrichment analysis Over representation analysis 1 Gene set enrichment analysis 1. Given an a priori defined set of genes S. 2. Rank genes (e.g. by t‐value between 2 groups of microarray samples) ranked gene list L. 3. Calculation of an enrichment score (ES) that reflects the degree to which a set S is overrepresented at the extremes (top or bottom) of the entire ranked list L. 4. Estimation the statistical significance (nominal P value) of the ES by using an empirical phenotype‐based permutation test procedure. 5. Adjustment for multiple hypothesis testing by controlling the false discovery rate (FDR). Gene set enrichment analysis Subramanian A et al. Proc Natl Acad Sci (2005) 2 Biochemical and Metabolic Pathways Böhringer Mannheim Signaling networks (in cancer cell) Hannah, Weinberg, Cell. 2000 137 NCI curated pathway maps (http://pid.nci.nih.gov) Signal Transduction Knowledge Environment http://stke.sciencemag.org/cm/ 3 Pathways • Pathways are available mechanistic information •Kyoto Encylopedia of Genes and Genomes, KEGG (http://www.genome.jp/kegg/) BioCYc • EcoCYC Metabolic Pathway Database (E. Coli) •Also for other organisms (e.g. HumanCYC) 4 Pathways • Pathways from Biocarta • http://www.biocarta.com/genes/index.asp Transpath Part of larger BioBase package (commercial) • PathwayBuilder package -

Nicotinic Signalling and Neurosteroid Modulation in Principal Neurons of the Hippocampal Formation and Prefrontal Cortex
Nicotinic Signalling and Neurosteroid Modulation in Principal Neurons of the Hippocampal Formation and Prefrontal Cortex by Beryl Yik Ting Chung A Thesis presented to The University of Guelph In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Sciences and Neuroscience Guelph, Ontario, Canada © Beryl Yik Ting Chung, April, 2018 ABSTRACT NICOTINIC SIGNALLING AND NEUROSTEROID MODULATION IN PRINCIPAL NEURONS OF THE HIPPOCAMPAL FORMATION AND PREFRONTAL CORTEX Beryl Yik Ting Chung Advisor: University of Guelph, 2018 Dr. Craig D.C. Bailey Nicotinic signalling plays an important role in coordinating the response of neuronal networks in many brain regions. During pre- and postnatal circuit formation, neurotransmission mediated by nicotinic acetylcholine receptors (nAChRs) influences neuronal survival and regulates neuronal excitability, synaptic transmission, and synaptic plasticity. Nicotinic signalling is also necessary for the proper function of the hippocampal formation (HF) and prefrontal cortex (PFC), which are anatomically and functionally connected and facilitate higher-order cognitive functions. The decline or dysfunction in nicotinic signalling and nAChR function has been observed in various neurological disorders, and the disruption or alteration of nicotinic signalling in the HF and/or PFC can impair learning and memory. While the location and functional role of the α4β2* nAChR isoform has been well characterized in the medial portion of the PFC, this is not well-established in the HF. What is the role of α4β2* nAChRs in excitatory principal neurons of the HF during early development? Growing evidence suggests that the progesterone metabolite allopregnanolone (ALLO) plays a role in mediating the proper function of the HF and the PFC, and that it may also inhibit nAChR function. -

Urinary Tract Effects of HPSE2 Mutations
BRIEF COMMUNICATION www.jasn.org Urinary Tract Effects of HPSE2 Mutations † Helen M. Stuart,* Neil A. Roberts,* Emma N. Hilton,* Edward A. McKenzie, Sarah B. Daly,* † ‡ Kristen D. Hadfield,* Jeffery S. Rahal,* Natalie J. Gardiner, Simon W. Tanley, | Malcolm A. Lewis,* Emily Sites,§ Brad Angle,§ Cláudia Alves, Teresa Lourenço,¶ Márcia Rodrigues,¶ Angelina Calado,** Marta Amado,** Nancy Guerreiro,** Inês Serras,** †† †† || Christian Beetz , Rita-Eva Varga , Mesrur Selcuk Silay,§§ John M. Darlow, ¶¶ || || ††† Mark G. Dobson , ¶¶ David E. Barton , *** Manuela Hunziker,¶¶ Prem Puri,¶¶*** ‡‡‡ Sally A. Feather, Judith A. Goodship ,§§§ Timothy H.J. Goodship ,§§§ Heather J. ||| BRIEF COMMUNICATION Lambert ,§§§ Heather J. Cordell ,§§§ the UK VUR Study Group, Anand Saggar, †††† Maria Kinali ,¶¶¶, the 4C Study Group, Christian Lorenz ,**** Kristina Moeller, |||| Franz Schaefer,§§§§ Aysun K. Bayazit , Stefanie Weber,¶¶¶¶ William G. Newman ,* and Adrian S. Woolf * Due to the number of contributing authors, the affiliations are listed at the end of this article. ABSTRACT Urofacial syndrome (UFS) is an autosomal recessive congenital disease featuring grimacing grimacing and dysmorphic bladders. and incomplete bladder emptying. Mutations of HPSE2, encoding heparanase 2, a Considering these and previously pub- heparanase 1 inhibitor, occur in UFS, but knowledge about the HPSE2 mutation spec- lished mutations4–7 (Figure 1A), it is clear trum is limited. Here, seven UFS kindreds with HPSE2 mutations are presented, that pathogenic HPSE2 mutations are including one with deleted asparagine 254, suggesting a role for this amino acid, found across the gene’s coding region. which is conserved in vertebrate orthologs. HPSE2 mutations were absent in 23 non- Most (i.e., nonsense or frameshift muta- neurogenic neurogenic bladder probands and, of 439 families with nonsyndromic tions) would cause loss of function, but a vesicoureteric reflux, only one carried a putative pathogenic HPSE2 variant. -

UNIVERSITY of CALIFORNIA, SAN DIEGO Functional Analysis of Sall4
UNIVERSITY OF CALIFORNIA, SAN DIEGO Functional analysis of Sall4 in modulating embryonic stem cell fate A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Molecular Pathology by Pei Jen A. Lee Committee in charge: Professor Steven Briggs, Chair Professor Geoff Rosenfeld, Co-Chair Professor Alexander Hoffmann Professor Randall Johnson Professor Mark Mercola 2009 Copyright Pei Jen A. Lee, 2009 All rights reserved. The dissertation of Pei Jen A. Lee is approved, and it is acceptable in quality and form for publication on microfilm and electronically: ______________________________________________________________ ______________________________________________________________ ______________________________________________________________ ______________________________________________________________ Co-Chair ______________________________________________________________ Chair University of California, San Diego 2009 iii Dedicated to my parents, my brother ,and my husband for their love and support iv Table of Contents Signature Page……………………………………………………………………….…iii Dedication…...…………………………………………………………………………..iv Table of Contents……………………………………………………………………….v List of Figures…………………………………………………………………………...vi List of Tables………………………………………………….………………………...ix Curriculum vitae…………………………………………………………………………x Acknowledgement………………………………………………….……….……..…...xi Abstract………………………………………………………………..…………….....xiii Chapter 1 Introduction ..…………………………………………………………………………….1 Chapter 2 Materials and Methods……………………………………………………………..…12 -

By Submitted in Partial Satisfaction of the Requirements for Degree of in In
Developments of Two Imaging based Technologies for Cell Biology Researches by Xiaowei Yan DISSERTATION Submitted in partial satisfaction of the requirements for degree of DOCTOR OF PHILOSOPHY in Biochemistry and Molecular Biology in the GRADUATE DIVISION of the UNIVERSITY OF CALIFORNIA, SAN FRANCISCO Approved: ______________________________________________________________________________Ronald Vale Chair ______________________________________________________________________________Jonathan Weissman ______________________________________________________________________________Orion Weiner ______________________________________________________________________________ ______________________________________________________________________________ Committee Members Copyright 2021 By Xiaowei Yan ii DEDICATION Everything happens for the best. To my family, who supported me with all their love. iii ACKNOWLEDGEMENTS The greatest joy of my PhD has been joining UCSF, working and learning with such a fantastic group of scientists. I am extremely grateful for all the support and mentorship I received and would like to thank: My mentor, Ron Vale, who is such a great and generous person. Thank you for showing me that science is so much fun and thank you for always giving me the freedom in pursuing my interest. I am grateful for all the guidance from you and thank you for always supporting me whenever I needed. You are a person full of wisdom, and I have been learning so much from you and your attitude to science, science community and even life will continue inspire me. Thank you for being my mentor and thank you for being such a great mentor. Everyone else in Vale lab, past and present, for making our lab a sweet home. I would like to give my special thank to Marvin (Marvin Tanenbaum) and Nico (Nico Stuurman), two other mentors for me in the lab. I would like to thank them for helping me adapt to our lab, for all the valuable advice and for all the happiness during the time that we work together. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
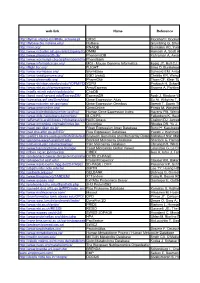
Web-Link Name Reference DRSC Flockhart I, Booker M, Kiger A, Et Al.: Flyrnai: the Drosophila Rnai Screening Center Database
web-link Name Reference http://flyrnai.org/cgi-bin/RNAi_screens.pl DRSC Flockhart I, Booker M, Kiger A, et al.: FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 34(Database issue):D489-94 (2006) http://flybase.bio.indiana.edu/ FlyBase Grumbling G, Strelets V.: FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34(Database issue):D484-8 (2006) http://rnai.org/ RNAiDB Gunsalus KC, Yueh WC, MacMenamin P, et al.: RNAiDB and PhenoBlast: web tools for genome-wide phenotypic mapping projects. Nucleic Acids Res. 32(Database issue):D406-10 (2004) http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIMOMIM Hamosh A, Scott AF, Amberger JS, et al.: Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33(Database issue):D514-7 (2005) http://www.phenomicdb.de/ PhenomicDB Kahraman A, Avramov A, Nashev LG, et al.: PhenomicDB: a multi-species genotype/phenotype database for comparative phenomics. Bioinformatics 21(3):418-20 (2005) http://www.worm.mpi-cbg.de/phenobank2/cgi-bin/MenuPage.pyPhenoBank http://www.informatics.jax.org/ MGI - Mouse Genome Informatics Eppig JT, Bult CJ, Kadin JA, et al.: The Mouse Genome Database (MGD): from genes to mice--a community resource for mouse biology. Nucleic Acids Res. 33(Database issue):D471-5 (2005) http://flight.licr.org/ FLIGHT Sims D, Bursteinas B, Gao Q, et al.: FLIGHT: database and tools for the integration and cross-correlation of large-scale RNAi phenotypic datasets. Nucleic Acids Res. 34(Database issue):D479-83 (2006) http://www.wormbase.org/ WormBase Schwarz EM, Antoshechkin I, Bastiani C, et al.: WormBase: better software, richer content. -

Epigenetic Mechanisms of Lncrnas Binding to Protein in Carcinogenesis
cancers Review Epigenetic Mechanisms of LncRNAs Binding to Protein in Carcinogenesis Tae-Jin Shin, Kang-Hoon Lee and Je-Yoel Cho * Department of Biochemistry, BK21 Plus and Research Institute for Veterinary Science, School of Veterinary Medicine, Seoul National University, Seoul 08826, Korea; [email protected] (T.-J.S.); [email protected] (K.-H.L.) * Correspondence: [email protected]; Tel.: +82-02-800-1268 Received: 21 September 2020; Accepted: 9 October 2020; Published: 11 October 2020 Simple Summary: The functional analysis of lncRNA, which has recently been investigated in various fields of biological research, is critical to understanding the delicate control of cells and the occurrence of diseases. The interaction between proteins and lncRNA, which has been found to be a major mechanism, has been reported to play an important role in cancer development and progress. This review thus organized the lncRNAs and related proteins involved in the cancer process, from carcinogenesis to metastasis and resistance to chemotherapy, to better understand cancer and to further develop new treatments for it. This will provide a new perspective on clinical cancer diagnosis, prognosis, and treatment. Abstract: Epigenetic dysregulation is an important feature for cancer initiation and progression. Long non-coding RNAs (lncRNAs) are transcripts that stably present as RNA forms with no translated protein and have lengths larger than 200 nucleotides. LncRNA can epigenetically regulate either oncogenes or tumor suppressor genes. Nowadays, the combined research of lncRNA plus protein analysis is gaining more attention. LncRNA controls gene expression directly by binding to transcription factors of target genes and indirectly by complexing with other proteins to bind to target proteins and cause protein degradation, reduced protein stability, or interference with the binding of other proteins.