By Submitted in Partial Satisfaction of the Requirements for Degree of in In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

PARSANA-DISSERTATION-2020.Pdf
DECIPHERING TRANSCRIPTIONAL PATTERNS OF GENE REGULATION: A COMPUTATIONAL APPROACH by Princy Parsana A dissertation submitted to The Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland July, 2020 © 2020 Princy Parsana All rights reserved Abstract With rapid advancements in sequencing technology, we now have the ability to sequence the entire human genome, and to quantify expression of tens of thousands of genes from hundreds of individuals. This provides an extraordinary opportunity to learn phenotype relevant genomic patterns that can improve our understanding of molecular and cellular processes underlying a trait. The high dimensional nature of genomic data presents a range of computational and statistical challenges. This dissertation presents a compilation of projects that were driven by the motivation to efficiently capture gene regulatory patterns in the human transcriptome, while addressing statistical and computational challenges that accompany this data. We attempt to address two major difficulties in this domain: a) artifacts and noise in transcriptomic data, andb) limited statistical power. First, we present our work on investigating the effect of artifactual variation in gene expression data and its impact on trans-eQTL discovery. Here we performed an in-depth analysis of diverse pre-recorded covariates and latent confounders to understand their contribution to heterogeneity in gene expression measurements. Next, we discovered 673 trans-eQTLs across 16 human tissues using v6 data from the Genotype Tissue Expression (GTEx) project. Finally, we characterized two trait-associated trans-eQTLs; one in Skeletal Muscle and another in Thyroid. Second, we present a principal component based residualization method to correct gene expression measurements prior to reconstruction of co-expression networks. -

Vps4b Heterozygous Mice Do Not Develop Tooth Defects That Replicate
Hu et al. BMC Genetics (2019) 20:7 https://doi.org/10.1186/s12863-018-0699-3 RESEARCH ARTICLE Open Access Vps4b heterozygous mice do not develop tooth defects that replicate human dentin dysplasia I Aiqin Hu1, Ting Lu1,2, Danna Chen1, Jin Huang1, Weiwei Feng1, Yanjun Li1, Dan Guo1, Xiangmin Xu1,3, Dong Chen1,4* and Fu Xiong1,3* Abstract Background: Vacuolar protein sorting-associated protein 4B (VPS4B) is a member of the ATP enzyme AAA protein family, and is mainly involved in protein degradation and cell membrane fusion. Recently, a dominant mutation in this gene was identified in human dentin dysplasia type I (DD-I). Herein, we report the generation of Vps4b knockout (Vps4b KO) mice; however, the homozygous Vps4b KO mutation was embryonic lethal at the early stages of embryo development, and we therefore report the results of heterozygous mutant mice. Results: Mice heterozygous for Vps4b did not develop tooth defects replicating human DD-I. Immunohistochemistry showed that gene KO was successful, as there was decreased expression of Vps4b in heterozygous mice; hematoxylin and eosin (H&E) staining also showed that the width of the pre-dentin zone was increased in heterozygous mice, although the arrangement of the odontoblasts was not significantly different from wild-type (WT) mice. However, H&E staining showed no obvious abnormalities in the bones of heterozygous mice. Moreover, stereomicroscopic and X-ray radiography results indicated no abnormal manifestations in teeth or bones. Furthermore, statistical analysis of the volume and density of dentin and enamel, as well as skeletal analysis, including the volume and separation of trabecular bone analyzed by micro-CT, all showed no differences between Vps4b heterozygotes and WT mice. -

Mouse Casp8ap2 Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Casp8ap2 Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Casp8ap2 conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Casp8ap2 gene (NCBI Reference Sequence: NM_011997 ; Ensembl: ENSMUSG00000028282 ) is located on Mouse chromosome 4. 11 exons are identified, with the ATG start codon in exon 2 and the TAG stop codon in exon 10 (Transcript: ENSMUST00000029950). Exon 4 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Casp8ap2 gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP23-75H14 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mice homozygous for disruption of this gene die before implantation. Exon 4 starts from about 2.12% of the coding region. The knockout of Exon 4 will result in frameshift of the gene. The size of intron 3 for 5'-loxP site insertion: 819 bp, and the size of intron 4 for 3'-loxP site insertion: 639 bp. The size of effective cKO region: ~601 bp. The cKO region does not have any other known gene. Page 1 of 7 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele gRNA region 5' gRNA region 3' 1 3 4 5 11 Targeting vector Targeted allele Constitutive KO allele (After Cre recombination) Legends Exon of mouse Casp8ap2 Homology arm cKO region loxP site Page 2 of 7 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Cytotoxic Effects and Changes in Gene Expression Profile
Toxicology in Vitro 34 (2016) 309–320 Contents lists available at ScienceDirect Toxicology in Vitro journal homepage: www.elsevier.com/locate/toxinvit Fusarium mycotoxin enniatin B: Cytotoxic effects and changes in gene expression profile Martina Jonsson a,⁎,MarikaJestoib, Minna Anthoni a, Annikki Welling a, Iida Loivamaa a, Ville Hallikainen c, Matti Kankainen d, Erik Lysøe e, Pertti Koivisto a, Kimmo Peltonen a,f a Chemistry and Toxicology Research Unit, Finnish Food Safety Authority (Evira), Mustialankatu 3, FI-00790 Helsinki, Finland b Product Safety Unit, Finnish Food Safety Authority (Evira), Mustialankatu 3, FI-00790 Helsinki, c The Finnish Forest Research Institute, Rovaniemi Unit, P.O. Box 16, FI-96301 Rovaniemi, Finland d Institute for Molecular Medicine Finland (FIMM), University of Helsinki, P.O. Box 20, FI-00014, Finland e Plant Health and Biotechnology, Norwegian Institute of Bioeconomy, Høyskoleveien 7, NO -1430 Ås, Norway f Finnish Safety and Chemicals Agency (Tukes), Opastinsilta 12 B, FI-00521 Helsinki, Finland article info abstract Article history: The mycotoxin enniatin B, a cyclic hexadepsipeptide produced by the plant pathogen Fusarium,isprevalentin Received 3 December 2015 grains and grain-based products in different geographical areas. Although enniatins have not been associated Received in revised form 5 April 2016 with toxic outbreaks, they have caused toxicity in vitro in several cell lines. In this study, the cytotoxic effects Accepted 28 April 2016 of enniatin B were assessed in relation to cellular energy metabolism, cell proliferation, and the induction of ap- Available online 6 May 2016 optosis in Balb 3T3 and HepG2 cells. The mechanism of toxicity was examined by means of whole genome ex- fi Keywords: pression pro ling of exposed rat primary hepatocytes. -

Epigenetic Mechanisms of Lncrnas Binding to Protein in Carcinogenesis
cancers Review Epigenetic Mechanisms of LncRNAs Binding to Protein in Carcinogenesis Tae-Jin Shin, Kang-Hoon Lee and Je-Yoel Cho * Department of Biochemistry, BK21 Plus and Research Institute for Veterinary Science, School of Veterinary Medicine, Seoul National University, Seoul 08826, Korea; [email protected] (T.-J.S.); [email protected] (K.-H.L.) * Correspondence: [email protected]; Tel.: +82-02-800-1268 Received: 21 September 2020; Accepted: 9 October 2020; Published: 11 October 2020 Simple Summary: The functional analysis of lncRNA, which has recently been investigated in various fields of biological research, is critical to understanding the delicate control of cells and the occurrence of diseases. The interaction between proteins and lncRNA, which has been found to be a major mechanism, has been reported to play an important role in cancer development and progress. This review thus organized the lncRNAs and related proteins involved in the cancer process, from carcinogenesis to metastasis and resistance to chemotherapy, to better understand cancer and to further develop new treatments for it. This will provide a new perspective on clinical cancer diagnosis, prognosis, and treatment. Abstract: Epigenetic dysregulation is an important feature for cancer initiation and progression. Long non-coding RNAs (lncRNAs) are transcripts that stably present as RNA forms with no translated protein and have lengths larger than 200 nucleotides. LncRNA can epigenetically regulate either oncogenes or tumor suppressor genes. Nowadays, the combined research of lncRNA plus protein analysis is gaining more attention. LncRNA controls gene expression directly by binding to transcription factors of target genes and indirectly by complexing with other proteins to bind to target proteins and cause protein degradation, reduced protein stability, or interference with the binding of other proteins. -
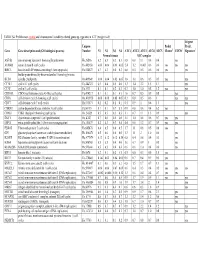
TABLE S4: Proliferation-Related and Chromosomal Instability-Related
TABLE S4: Proliferation-related and chromosomal instability-related genes up-regulated in ATC (weight <4.0) 44-gene Unigene Prolif. Prolif. Gene Gene description and (GO biological process) Number N1 N2 N3 N4 ATC1 ATC2 ATC3 ATC4 ATC5 Cluster1 CIN702 Signature3 Normal tissues ATC samples ASF1B anti-silencing function 1 homolog B (unknown) Hs.26516 0.2 0.3 0.3 0.3 0.8 0.8 1.1 0.8 0.4 yes AURKB aurora kinase B (cell cycle) Hs.442658 0.06 0.06 0.08 0.05 2.4 1.2 0.003 0.6 0.6 yes yes yes BIRC5 baculoviral IAP repeat-containing 5 (anti apoptosis) Hs.514527 0.7 0.3 0.4 0.3 0.8 0.5 0.5 0.6 0.6 yes yes budding uninhibited by benzimidazoles 1 homolog (mitotic BUB1 spindle checkpoint) Hs.469649 0.06 0.04 0.02 0.05 1.8 1.0 0.6 0.5 0.5 yes yes CCNE1 cyclin E1 (cell cycle) Hs.244723 0.5 0.4 0.5 0.6 1.3 1.4 2.2 1.5 1.1 yes CCNF cyclin F (cell cycle) Hs.1973 0.1 0.1 0.2 0.3 0.7 1.0 1.0 0.8 1.2 yes yes CDC45L CDC45 cell division cycle 45-like (cell cycle) Hs.474217 0.1 0.1 0.1 0.3 1.6 0.7 0.5 0.9 0.8 yes CDC6 cell division cycle 6 homolog (cell cycle) Hs. 405958 0.08 0.08 0.08 0.07 0.3 0.8 0.5 0.6 1 yes yes CDC7 cell division cycle 7 (cell cycle) Hs.533573 0.2 0.2 0.2 0.1 0.5 0.9 1 0.6 1.3 yes CDKN3 cyclin-dependent kinase inhibitor 3 (cell cycle) Hs.84113 0.1 0.1 0.7 0.1 0.9 0.8 0.6 0.4 6.2 yes CHEK1 CHK1 checkpoint homolog (cell cycle) Hs. -

1 Supporting Information for a Microrna Network Regulates
Supporting Information for A microRNA Network Regulates Expression and Biosynthesis of CFTR and CFTR-ΔF508 Shyam Ramachandrana,b, Philip H. Karpc, Peng Jiangc, Lynda S. Ostedgaardc, Amy E. Walza, John T. Fishere, Shaf Keshavjeeh, Kim A. Lennoxi, Ashley M. Jacobii, Scott D. Rosei, Mark A. Behlkei, Michael J. Welshb,c,d,g, Yi Xingb,c,f, Paul B. McCray Jr.a,b,c Author Affiliations: Department of Pediatricsa, Interdisciplinary Program in Geneticsb, Departments of Internal Medicinec, Molecular Physiology and Biophysicsd, Anatomy and Cell Biologye, Biomedical Engineeringf, Howard Hughes Medical Instituteg, Carver College of Medicine, University of Iowa, Iowa City, IA-52242 Division of Thoracic Surgeryh, Toronto General Hospital, University Health Network, University of Toronto, Toronto, Canada-M5G 2C4 Integrated DNA Technologiesi, Coralville, IA-52241 To whom correspondence should be addressed: Email: [email protected] (M.J.W.); yi- [email protected] (Y.X.); Email: [email protected] (P.B.M.) This PDF file includes: Materials and Methods References Fig. S1. miR-138 regulates SIN3A in a dose-dependent and site-specific manner. Fig. S2. miR-138 regulates endogenous SIN3A protein expression. Fig. S3. miR-138 regulates endogenous CFTR protein expression in Calu-3 cells. Fig. S4. miR-138 regulates endogenous CFTR protein expression in primary human airway epithelia. Fig. S5. miR-138 regulates CFTR expression in HeLa cells. Fig. S6. miR-138 regulates CFTR expression in HEK293T cells. Fig. S7. HeLa cells exhibit CFTR channel activity. Fig. S8. miR-138 improves CFTR processing. Fig. S9. miR-138 improves CFTR-ΔF508 processing. Fig. S10. SIN3A inhibition yields partial rescue of Cl- transport in CF epithelia. -

XIAO-DISSERTATION-2015.Pdf
CELLULAR AND PROCESS ENGINEERING TO IMPROVE MAMMALIAN MEMBRANE PROTEIN EXPRESSION By Su Xiao A dissertation is submitted to Johns Hopkins University in conformity with the requirements for degree of Doctor of Philosophy Baltimore, Maryland May 2015 © 2015 Su Xiao All Rights Reserved Abstract Improving the expression level of recombinant mammalian proteins has been pursued for production of commercial biotherapeutics in industry, as well as for biomedical studies in academia, as an adequate supply of correctly folded proteins is a prerequisite for all structure and function studies. Presented in this dissertation are different strategies to improve protein functional expression level, especially for membrane proteins. The model protein is neurotensin receptor 1 (NTSR1), a hard-to- express G protein-coupled receptor (GPCR). GPCRs are integral membrane proteins playing a central role in cell signaling and are targets for most of the medicines sold worldwide. Obtaining adequate functional GPCRs has been a bottleneck in their structure studies because the expression of these proteins from mammalian cells is very low. The first strategy is the adoption of mammalian inducible expression system. A stable and inducible T-REx-293 cell line overexpressing an engineered rat NTSR1 was constructed. 2.5 million Functional copies of NTSR1 per cell were detected on plasma membrane, which is 167 fold improvement comparing to NTSR1 constitutive expression. The second strategy is production process development including suspension culture adaptation and induction parameter optimization. A further 3.5 fold improvement was achieved and approximately 1 milligram of purified functional NTSR1 per liter suspension culture was obtained. This was comparable yield to the transient baculovirus- insect cell system. -

1 AGING Supplementary Table 2
SUPPLEMENTARY TABLES Supplementary Table 1. Details of the eight domain chains of KIAA0101. Serial IDENTITY MAX IN COMP- INTERFACE ID POSITION RESOLUTION EXPERIMENT TYPE number START STOP SCORE IDENTITY LEX WITH CAVITY A 4D2G_D 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ B 4D2G_E 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ C 6EHT_D 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ D 6EHT_E 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ E 6GWS_D 41-72 41 72 100 100 3.2Å PCNA X-RAY DIFFRACTION √ F 6GWS_E 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ G 6GWS_F 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ H 6IIW_B 2-11 2 11 100 100 1.699Å UHRF1 X-RAY DIFFRACTION √ www.aging-us.com 1 AGING Supplementary Table 2. Significantly enriched gene ontology (GO) annotations (cellular components) of KIAA0101 in lung adenocarcinoma (LinkedOmics). Leading Description FDR Leading Edge Gene EdgeNum RAD51, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, CENPW, HJURP, NDC80, CDCA5, NCAPH, BUB1, ZWILCH, CENPK, KIF2C, AURKA, CENPN, TOP2A, CENPM, PLK1, ERCC6L, CDT1, CHEK1, SPAG5, CENPH, condensed 66 0 SPC24, NUP37, BLM, CENPE, BUB3, CDK2, FANCD2, CENPO, CENPF, BRCA1, DSN1, chromosome MKI67, NCAPG2, H2AFX, HMGB2, SUV39H1, CBX3, TUBG1, KNTC1, PPP1CC, SMC2, BANF1, NCAPD2, SKA2, NUP107, BRCA2, NUP85, ITGB3BP, SYCE2, TOPBP1, DMC1, SMC4, INCENP. RAD51, OIP5, CDK1, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, ESCO2, CENPW, HJURP, TTK, NDC80, CDCA5, BUB1, ZWILCH, CENPK, KIF2C, AURKA, DSCC1, CENPN, CDCA8, CENPM, PLK1, MCM6, ERCC6L, CDT1, HELLS, CHEK1, SPAG5, CENPH, PCNA, SPC24, CENPI, NUP37, FEN1, chromosomal 94 0 CENPL, BLM, KIF18A, CENPE, MCM4, BUB3, SUV39H2, MCM2, CDK2, PIF1, DNA2, region CENPO, CENPF, CHEK2, DSN1, H2AFX, MCM7, SUV39H1, MTBP, CBX3, RECQL4, KNTC1, PPP1CC, CENPP, CENPQ, PTGES3, NCAPD2, DYNLL1, SKA2, HAT1, NUP107, MCM5, MCM3, MSH2, BRCA2, NUP85, SSB, ITGB3BP, DMC1, INCENP, THOC3, XPO1, APEX1, XRCC5, KIF22, DCLRE1A, SEH1L, XRCC3, NSMCE2, RAD21. -

USP16 Counteracts Mono-Ubiquitination of Rps27a And
RESEARCH ARTICLE USP16 counteracts mono-ubiquitination of RPS27a and promotes maturation of the 40S ribosomal subunit Christian Montellese1†, Jasmin van den Heuvel1,2, Caroline Ashiono1, Kerstin Do¨ rner1,2, Andre´ Melnik3‡, Stefanie Jonas1§, Ivo Zemp1, Paola Picotti3, Ludovic C Gillet1, Ulrike Kutay1* 1Institute of Biochemistry, ETH Zurich, Zurich, Switzerland; 2Molecular Life Sciences Ph.D. Program, Zurich, Switzerland; 3Institute of Molecular Systems Biology, ETH Zurich, Zurich, Switzerland Abstract Establishment of translational competence represents a decisive cytoplasmic step in the biogenesis of 40S ribosomal subunits. This involves final 18S rRNA processing and release of residual biogenesis factors, including the protein kinase RIOK1. To identify novel proteins promoting the final maturation of human 40S subunits, we characterized pre-ribosomal subunits trapped on RIOK1 by mass spectrometry, and identified the deubiquitinase USP16 among the captured factors. We demonstrate that USP16 constitutes a component of late cytoplasmic pre-40S subunits that promotes the removal of ubiquitin from an internal lysine of ribosomal protein *For correspondence: RPS27a/eS31. USP16 deletion leads to late 40S subunit maturation defects, manifesting in [email protected] incomplete processing of 18S rRNA and retarded recycling of late-acting ribosome biogenesis Present address: †CSL Behring, factors, revealing an unexpected contribution of USP16 to the ultimate step of 40S synthesis. CSL Biologics Research Center, Finally, ubiquitination of RPS27a appears to depend on active translation, pointing at a potential ‡ Bern, Switzerland; MSD Merck connection between 40S maturation and protein synthesis. Sharp & Dohme AG, Lucerne, Switzerland; §Institute of Molecular Biology and Biophysics, ETH Zurich, Zurich, Switzerland Introduction Ribosomes stand at the center of translation in all kingdoms of life, catalyzing the synthesis of pro- Competing interests: The teins by reading a messenger RNA (mRNA) template. -

Supplementary Data
SUPPLEMENTARY DATA A cyclin D1-dependent transcriptional program predicts clinical outcome in mantle cell lymphoma Santiago Demajo et al. 1 SUPPLEMENTARY DATA INDEX Supplementary Methods p. 3 Supplementary References p. 8 Supplementary Tables (S1 to S5) p. 9 Supplementary Figures (S1 to S15) p. 17 2 SUPPLEMENTARY METHODS Western blot, immunoprecipitation, and qRT-PCR Western blot (WB) analysis was performed as previously described (1), using cyclin D1 (Santa Cruz Biotechnology, sc-753, RRID:AB_2070433) and tubulin (Sigma-Aldrich, T5168, RRID:AB_477579) antibodies. Co-immunoprecipitation assays were performed as described before (2), using cyclin D1 antibody (Santa Cruz Biotechnology, sc-8396, RRID:AB_627344) or control IgG (Santa Cruz Biotechnology, sc-2025, RRID:AB_737182) followed by protein G- magnetic beads (Invitrogen) incubation and elution with Glycine 100mM pH=2.5. Co-IP experiments were performed within five weeks after cell thawing. Cyclin D1 (Santa Cruz Biotechnology, sc-753), E2F4 (Bethyl, A302-134A, RRID:AB_1720353), FOXM1 (Santa Cruz Biotechnology, sc-502, RRID:AB_631523), and CBP (Santa Cruz Biotechnology, sc-7300, RRID:AB_626817) antibodies were used for WB detection. In figure 1A and supplementary figure S2A, the same blot was probed with cyclin D1 and tubulin antibodies by cutting the membrane. In figure 2H, cyclin D1 and CBP blots correspond to the same membrane while E2F4 and FOXM1 blots correspond to an independent membrane. Image acquisition was performed with ImageQuant LAS 4000 mini (GE Healthcare). Image processing and quantification were performed with Multi Gauge software (Fujifilm). For qRT-PCR analysis, cDNA was generated from 1 µg RNA with qScript cDNA Synthesis kit (Quantabio). qRT–PCR reaction was performed using SYBR green (Roche). -

Role of Senataxin in RNA: DNA Hybrids Resolution at DNA Double
Role of senataxin in RNA : DNA hybrids resolution at DNA double strand breaks Sarah Cohen To cite this version: Sarah Cohen. Role of senataxin in RNA : DNA hybrids resolution at DNA double strand breaks. Cellular Biology. Université Paul Sabatier - Toulouse III, 2019. English. NNT : 2019TOU30125. tel-02930730 HAL Id: tel-02930730 https://tel.archives-ouvertes.fr/tel-02930730 Submitted on 4 Sep 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. ����� ���� ������������������ ������������������������������������ ��������� ���!������"������ �� "��#�$�% ��� � ���� %��"���������"� ��� ��� � � � &������ ������������ ������� ����'������"�� � (����� �"�� ���"�� �������"�&)�����"����*���� (�+ "" ��"��� ��������"�����!��� ������������������,�,�$�,����-��.�� ���.�,����+&����-��" �������������,����/�������������� �������������������� �,�0�%�$�� ��� ���������,����-������� � �������0���+ � ����� ������'������� ������'������������ %����1�� ����� ����������������� /��������/�,�� ��� ���������� ������������������� ������������������������������ �� �����������������