Inference for Health Econometrics: Inference, Model Tests, Diagnostics, Multiple Tests, and Bootstrap
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

BIOGRAPHICAL SKETCH Garrett M. Fitzmaurice Associate Professor Of
Principal Investigator/Program Director (Last, First, Middle): BIOGRAPHICAL SKETCH Provide the following information for the key personnel and other significant contributors in the order listed on Form Page 2. Follow this format for each person. DO NOT EXCEED FOUR PAGES. NAME POSITION TITLE Garrett M. Fitzmaurice Associate Professor of Biostatistics eRA COMMONS USER NAME EDUCATION/TRAINING (Begin with baccalaureate or other initial professional education, such as nursing, and include postdoctoral training.) DEGREE INSTITUTION AND LOCATION YEAR(s) FIELD OF STUDY (if applicable) National University of Ireland BA, MA 1983, 1987 Psychology University of London MSc 1986 Quantitative Methods Harvard University ScD 1993 Biostatistics Professional Experience: 1986-1989 Statistician, Department of Psychology, New York University 1989-1990 Teaching Assistant, Department of Biostatistics, Harvard School of Public Health 1990-1993 Teaching Fellow in Biostatistics, Department of Biostatistics, Harvard SPH 1993-1994 Post-doctoral Research Fellow, Department of Biostatistics, Harvard SPH 1994-1997 Research Fellow, Nuffield College, Oxford University, United Kingdom 1997-1999 Assistant Professor of Biostatistics, Harvard School of Public Health 1999-present Associate Professor of Biostatistics, Harvard School of Public Health 2004-present Associate Professor of Medicine (Biostatistics), Harvard Medical School 2004-present Biostatistician, Division of General Medicine, Brigham and Women’s Hospital, Boston 2006-present Foreign Adjunct Professor of Biostatistics, -

TUTORIAL in BIOSTATISTICS: the Self-Controlled Case Series Method
STATISTICS IN MEDICINE Statist. Med. 2005; 0:1–31 Prepared using simauth.cls [Version: 2002/09/18 v1.11] TUTORIAL IN BIOSTATISTICS: The self-controlled case series method Heather J. Whitaker1, C. Paddy Farrington1, Bart Spiessens2 and Patrick Musonda1 1 Department of Statistics, The Open University, Milton Keynes, MK7 6AA, UK. 2 GlaxoSmithKline Biologicals, Rue de l’Institut 89, B-1330 Rixensart, Belgium. SUMMARY The self-controlled case series method was developed to investigate associations between acute outcomes and transient exposures, using only data on cases, that is, on individuals who have experienced the outcome of interest. Inference is within individuals, and hence fixed covariates effects are implicitly controlled for within a proportional incidence framework. We describe the origins, assumptions, limitations, and uses of the method. The rationale for the model and the derivation of the likelihood are explained in detail using a worked example on vaccine safety. Code for fitting the model in the statistical package STATA is described. Two further vaccine safety data sets are used to illustrate a range of modelling issues and extensions of the basic model. Some brief pointers on the design of case series studies are provided. The data sets, STATA code, and further implementation details in SAS, GENSTAT and GLIM are available from an associated website. key words: case series; conditional likelihood; control; epidemiology; modelling; proportional incidence Copyright c 2005 John Wiley & Sons, Ltd. 1. Introduction The self-controlled case series method, or case series method for short, provides an alternative to more established cohort or case-control methods for investigating the association between a time-varying exposure and an outcome event. -

Biometrics & Biostatistics
Hanley and Moodie, J Biomet Biostat 2011, 2:5 Biometrics & Biostatistics http://dx.doi.org/10.4172/2155-6180.1000124 Research Article Article OpenOpen Access Access Sample Size, Precision and Power Calculations: A Unified Approach James A Hanley* and Erica EM Moodie Department of Epidemiology, Biostatistics, and Occupational Health, McGill University, Canada Abstract The sample size formulae given in elementary biostatistics textbooks deal only with simple situations: estimation of one, or a comparison of at most two, mean(s) or proportion(s). While many specialized textbooks give sample formulae/tables for analyses involving odds and rate ratios, few deal explicitly with statistical considera tions for slopes (regression coefficients), for analyses involving confounding variables or with the fact that most analyses rely on some type of generalized linear model. Thus, the investigator is typically forced to use “black-box” computer programs or tables, or to borrow from tables in the social sciences, where the emphasis is on cor- relation coefficients. The concern in the – usually very separate – modules or stand alone software programs is more with user friendly input and output. The emphasis on numerical exactness is particularly unfortunate, given the rough, prospective, and thus uncertain, nature of the exercise, and that different textbooks and software may give different sample sizes for the same design. In addition, some programs focus on required numbers per group, others on an overall number. We present users with a single universal (though sometimes approximate) formula that explicitly isolates the impacts of the various factors one from another, and gives some insight into the determinants for each factor. -

The American Statistician
This article was downloaded by: [T&F Internal Users], [Rob Calver] On: 01 September 2015, At: 02:24 Publisher: Taylor & Francis Informa Ltd Registered in England and Wales Registered Number: 1072954 Registered office: 5 Howick Place, London, SW1P 1WG The American Statistician Publication details, including instructions for authors and subscription information: http://www.tandfonline.com/loi/utas20 Reviews of Books and Teaching Materials Published online: 27 Aug 2015. Click for updates To cite this article: (2015) Reviews of Books and Teaching Materials, The American Statistician, 69:3, 244-252, DOI: 10.1080/00031305.2015.1068616 To link to this article: http://dx.doi.org/10.1080/00031305.2015.1068616 PLEASE SCROLL DOWN FOR ARTICLE Taylor & Francis makes every effort to ensure the accuracy of all the information (the “Content”) contained in the publications on our platform. However, Taylor & Francis, our agents, and our licensors make no representations or warranties whatsoever as to the accuracy, completeness, or suitability for any purpose of the Content. Any opinions and views expressed in this publication are the opinions and views of the authors, and are not the views of or endorsed by Taylor & Francis. The accuracy of the Content should not be relied upon and should be independently verified with primary sources of information. Taylor and Francis shall not be liable for any losses, actions, claims, proceedings, demands, costs, expenses, damages, and other liabilities whatsoever or howsoever caused arising directly or indirectly in connection with, in relation to or arising out of the use of the Content. This article may be used for research, teaching, and private study purposes. -
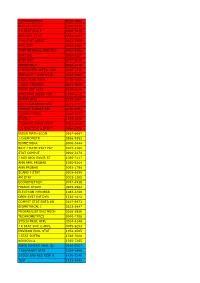
Econometrica 0012-9682 Biostatistics 1465-4644 J R
ECONOMETRICA 0012-9682 BIOSTATISTICS 1465-4644 J R STAT SOC B 1369-7412 ANN APPL STAT 1932-6157 J AM STAT ASSOC 0162-1459 ANN STAT 0090-5364 STAT METHODS MED RES 0962-2802 STAT SCI 0883-4237 STAT MED 0277-6715 BIOMETRICS 0006-341X CHEMOMETR INTELL LAB 0169-7439 IEEE ACM T COMPUT BI 1545-5963 J BUS ECON STAT 0735-0015 J QUAL TECHNOL 0022-4065 FUZZY SET SYST 0165-0114 STAT APPL GENET MOL 1544-6115 PHARM STAT 1539-1604 MULTIVAR BEHAV RES 0027-3171 PROBAB THEORY REL 0178-8051 J COMPUT BIOL 1066-5277 STATA J 1536-867X J COMPUT GRAPH STAT 1061-8600 J R STAT SOC A STAT 0964-1998 INSUR MATH ECON 0167-6687 J CHEMOMETR 0886-9383 BIOMETRIKA 0006-3444 BRIT J MATH STAT PSY 0007-1102 STAT COMPUT 0960-3174 J AGR BIOL ENVIR ST 1085-7117 ANN APPL PROBAB 1050-5164 ANN PROBAB 0091-1798 SCAND J STAT 0303-6898 AM STAT 0003-1305 ECONOMET REV 0747-4938 FINANC STOCH 0949-2984 ELECTRON J PROBAB 1083-6489 OPEN SYST INF DYN 1230-1612 COMPUT STAT DATA AN 0167-9473 BIOMETRICAL J 0323-3847 PROBABILIST ENG MECH 0266-8920 TECHNOMETRICS 0040-1706 STOCH PROC APPL 0304-4149 J R STAT SOC C-APPL 0035-9254 ENVIRON ECOL STAT 1352-8505 J STAT SOFTW 1548-7660 BERNOULLI 1350-7265 INFIN DIMENS ANAL QU 0219-0257 J BIOPHARM STAT 1054-3406 STOCH ENV RES RISK A 1436-3240 TEST 1133-0686 ASTIN BULL 0515-0361 ADV APPL PROBAB 0001-8678 J TIME SER ANAL 0143-9782 MATH POPUL STUD 0889-8480 COMB PROBAB COMPUT 0963-5483 LIFETIME DATA ANAL 1380-7870 METHODOL COMPUT APPL 1387-5841 ECONOMET J 1368-4221 STATISTICS 0233-1888 J APPL PROBAB 0021-9002 J MULTIVARIATE ANAL 0047-259X ENVIRONMETRICS 1180-4009 -

Curriculum Vitae Jianwen Cai
CURRICULUM VITAE JIANWEN CAI Business Address: Department of Biostatistics, CB # 7420 The University of North Carolina at Chapel Hill Chapel Hill, NC 27599-7420 TEL: (919) 966-7788 FAX: (919) 966-3804 Email: [email protected] Education: 1992 - Ph.D. (Biostatistics), University of Washington, Seattle, Washington. 1989 - M.S. (Biostatistics), University of Washington, Seattle, Washington. 1985 - B.S. (Mathematics), Shandong University, Jinan, P. R. China. Positions: Teaching Assistant: Department of Biostatistics, University of Washington, 1988 - 1990. Research Assistant: Cardiovascular Health Study, University of Washington, 1988 - 1990. Fred Hutchinson Cancer Research Center, 1990 - 1992. Research Associate: Fred Hutchinson Cancer Research Center, 1992 - 1992. Assistant Professor: Department of Biostatistics, University of North Carolina at Chapel Hill, 1992 - 1999. Associate Professor: Department of Biostatistics, University of North Carolina at Chapel Hill, 1999 - 2004. Professor: Department of Biostatistics, University of North Carolina at Chapel Hill, 2004 - present. Interim Chair: Department of Biostatistics, University of North Carolina at Chapel Hill, 2006. Associate Chair: Department of Biostatistics, University of North Carolina at Chapel Hill, 2006-present. Professional Societies: American Statistical Association (ASA) International Biometric Society (ENAR) Institute of Mathematical Statistics (IMS) International Chinese Statistical Association (ICSA) 1 Research Interests: Survival Analysis and Regression Models, Design and Analysis of Clinical Trials, Analysis of Correlated Responses, Cancer Biology, Cardiovascular Disease Research, Obesity Research, Epidemiological Models. Awards/Honors: UNC School of Public Health McGavran Award for Excellence in Teaching, 2004. American Statistical Association Fellow, 2005 Institute of Mathematical Statistics Fellow, 2009 Professional Service: Associate Editor: Biometrics: 2000-2010. Lifetime Data Analysis: 2002-. Statistics in Biosciences: 2009-. Board Member: ENAR Regional Advisory Board, 1996-1998. -

Ying Zhang, Phd ______
Ying Zhang, PhD ________________________________________________________________ Home and Campus Addresses 19204 Sahler St. Elkhorn, NE 68022 Department of Biostatistics College of Public Health University of Nebraska Medical Center 984375 Nebraska Medical Center, Omaha, NE 68198-4375 Phone: (402) 836-9380 Email: [email protected] Education BS Computational Mathematics, Fundan University, China 1985 MS Computational Mathematics, Fundan University, China 1988 MS Applied Mathematics, Florida State University 1994 PhD Statistics, University of Washington 1998 Academic Appointments 07/2019 – current Professor & Chair, Department of Biostatistics, University of Nebraska Medical Center 08/2020 – current Professor & Acting Chair, Department of Epidemiology, University of Nebraska Medical Center 01/2014 – 06/2019 Professor & Director of Graduate Education, Department of Biostatistics, Indiana University 07/2010 - 06/2014 Professor, Department of Biostatistics, Program of Health Informatics and Program of Applied Mathematics, The University of Iowa 07/2004 – 06/2010 Associate Professor, Department of Biostatistics, The University of Iowa College of Public Health 08/1998 – 06/2004 Assistant Professor, Department of Statistics and Actuarial Science, University of Central Florida 06/1997 – 09/1997 Applied Statistician, Applied Statistics Group, The Boeing Company 09/1988 – 07/1991 Lecturer, Department of Mathematics, Fudan University, China Honors and Awards 2018 Elected Fellow of the American Statistical Association (ASA) 2008 “Quality of Life Award” by the Oncology Nursing Society Press for the paper “Transition from Treatment to Survivorship: Effects of a Psychoeducational Intervention on Quality of Life in 1 Breast Cancer Survivors” 2008 “Faculty Teaching Award” by the College of Public Health, University of Iowa 1997 “Z.W. Birnbaum award for excellence in dissertation proposal”; awarded by the Department of Statistics, University of Washington, 10/1997. -

Adaptive Enrichment Designs in Clinical Trials
Annual Review of Statistics and Its Application Adaptive Enrichment Designs in Clinical Trials Peter F. Thall Department of Biostatistics, M.D. Anderson Cancer Center, University of Texas, Houston, Texas 77030, USA; email: [email protected] Annu. Rev. Stat. Appl. 2021. 8:393–411 Keywords The Annual Review of Statistics and Its Application is adaptive signature design, Bayesian design, biomarker, clinical trial, group online at statistics.annualreviews.org sequential design, precision medicine, subset selection, targeted therapy, https://doi.org/10.1146/annurev-statistics-040720- variable selection 032818 Copyright © 2021 by Annual Reviews. Abstract All rights reserved Adaptive enrichment designs for clinical trials may include rules that use in- terim data to identify treatment-sensitive patient subgroups, select or com- pare treatments, or change entry criteria. A common setting is a trial to Annu. Rev. Stat. Appl. 2021.8:393-411. Downloaded from www.annualreviews.org compare a new biologically targeted agent to standard therapy. An enrich- ment design’s structure depends on its goals, how it accounts for patient heterogeneity and treatment effects, and practical constraints. This article Access provided by University of Texas - M.D. Anderson Cancer Center on 03/10/21. For personal use only. first covers basic concepts, including treatment-biomarker interaction, pre- cision medicine, selection bias, and sequentially adaptive decision making, and briefly describes some different types of enrichment. Numerical illus- trations are provided for qualitatively different cases involving treatment- biomarker interactions. Reviews are given of adaptive signature designs; a Bayesian design that uses a random partition to identify treatment-sensitive biomarker subgroups and assign treatments; and designs that enrich superior treatment sample sizes overall or within subgroups, make subgroup-specific decisions, or include outcome-adaptive randomization. -

Rank Full Journal Title Journal Impact Factor 1 Journal of Statistical
Journal Data Filtered By: Selected JCR Year: 2019 Selected Editions: SCIE Selected Categories: 'STATISTICS & PROBABILITY' Selected Category Scheme: WoS Rank Full Journal Title Journal Impact Eigenfactor Total Cites Factor Score Journal of Statistical Software 1 25,372 13.642 0.053040 Annual Review of Statistics and Its Application 2 515 5.095 0.004250 ECONOMETRICA 3 35,846 3.992 0.040750 JOURNAL OF THE AMERICAN STATISTICAL ASSOCIATION 4 36,843 3.989 0.032370 JOURNAL OF THE ROYAL STATISTICAL SOCIETY SERIES B-STATISTICAL METHODOLOGY 5 25,492 3.965 0.018040 STATISTICAL SCIENCE 6 6,545 3.583 0.007500 R Journal 7 1,811 3.312 0.007320 FUZZY SETS AND SYSTEMS 8 17,605 3.305 0.008740 BIOSTATISTICS 9 4,048 3.098 0.006780 STATISTICS AND COMPUTING 10 4,519 3.035 0.011050 IEEE-ACM Transactions on Computational Biology and Bioinformatics 11 3,542 3.015 0.006930 JOURNAL OF BUSINESS & ECONOMIC STATISTICS 12 5,921 2.935 0.008680 CHEMOMETRICS AND INTELLIGENT LABORATORY SYSTEMS 13 9,421 2.895 0.007790 MULTIVARIATE BEHAVIORAL RESEARCH 14 7,112 2.750 0.007880 INTERNATIONAL STATISTICAL REVIEW 15 1,807 2.740 0.002560 Bayesian Analysis 16 2,000 2.696 0.006600 ANNALS OF STATISTICS 17 21,466 2.650 0.027080 PROBABILISTIC ENGINEERING MECHANICS 18 2,689 2.411 0.002430 BRITISH JOURNAL OF MATHEMATICAL & STATISTICAL PSYCHOLOGY 19 1,965 2.388 0.003480 ANNALS OF PROBABILITY 20 5,892 2.377 0.017230 STOCHASTIC ENVIRONMENTAL RESEARCH AND RISK ASSESSMENT 21 4,272 2.351 0.006810 JOURNAL OF COMPUTATIONAL AND GRAPHICAL STATISTICS 22 4,369 2.319 0.008900 STATISTICAL METHODS IN -

Abbreviations of Names of Serials
Abbreviations of Names of Serials This list gives the form of references used in Mathematical Reviews (MR). ∗ not previously listed The abbreviation is followed by the complete title, the place of publication x journal indexed cover-to-cover and other pertinent information. y monographic series Update date: January 30, 2018 4OR 4OR. A Quarterly Journal of Operations Research. Springer, Berlin. ISSN xActa Math. Appl. Sin. Engl. Ser. Acta Mathematicae Applicatae Sinica. English 1619-4500. Series. Springer, Heidelberg. ISSN 0168-9673. y 30o Col´oq.Bras. Mat. 30o Col´oquioBrasileiro de Matem´atica. [30th Brazilian xActa Math. Hungar. Acta Mathematica Hungarica. Akad. Kiad´o,Budapest. Mathematics Colloquium] Inst. Nac. Mat. Pura Apl. (IMPA), Rio de Janeiro. ISSN 0236-5294. y Aastaraam. Eesti Mat. Selts Aastaraamat. Eesti Matemaatika Selts. [Annual. xActa Math. Sci. Ser. A Chin. Ed. Acta Mathematica Scientia. Series A. Shuxue Estonian Mathematical Society] Eesti Mat. Selts, Tartu. ISSN 1406-4316. Wuli Xuebao. Chinese Edition. Kexue Chubanshe (Science Press), Beijing. ISSN y Abel Symp. Abel Symposia. Springer, Heidelberg. ISSN 2193-2808. 1003-3998. y Abh. Akad. Wiss. G¨ottingenNeue Folge Abhandlungen der Akademie der xActa Math. Sci. Ser. B Engl. Ed. Acta Mathematica Scientia. Series B. English Wissenschaften zu G¨ottingen.Neue Folge. [Papers of the Academy of Sciences Edition. Sci. Press Beijing, Beijing. ISSN 0252-9602. in G¨ottingen.New Series] De Gruyter/Akademie Forschung, Berlin. ISSN 0930- xActa Math. Sin. (Engl. Ser.) Acta Mathematica Sinica (English Series). 4304. Springer, Berlin. ISSN 1439-8516. y Abh. Akad. Wiss. Hamburg Abhandlungen der Akademie der Wissenschaften xActa Math. Sinica (Chin. Ser.) Acta Mathematica Sinica. -

Short Course of the 2019 Workshop on Biostatistics and Bioinformatics
Short Course of the 2019 Workshop on Biostatistics and Bioinformatics Title: Phase II Clinical Development of New Drugs Room 1441, 25 Park Place, Georgia State University, Atlanta, GA 30303 1:30pm-5:30pm, May 10, 2019 Naitee Ting, Biostatistics & Data Sciences, Boehringer and Ingelheim Pharmaceuticals Inc. Abstract: In the process of medicine discovery and development, understanding the dose-response relationship is one of the most important and challenging tasks. It is critical to identify the right range of doses in early stages of medicine development so that Phase III trials can be properly designed to confirm appropriate dose(s) for the patient. Usually in the beginning of Phase II development, there is limited information to help guide trial designs, therefore Phase II clinical trials often consists of objectives for establishing proof of concept (PoC), identifying a set of potentially safe and efficacious doses, and characterizing the dose-response relationship. Some of the major challenges in designing these Phase II trials include the selection of dose range and frequency, clinical endpoints and/or biomarkers, and the use of control(s). Inappropriate Phase II trial designs may lead to delay of the medicine development program or waste of investment. Specifically, misleading results from poorly designed Phase II trials could force a Phase III program to confirm sub-optimal dose(s), or even stop developing a potentially useful medicine. Therefore, it is critical to consider Phase II trial designs, in the broader context of the entire medicine development plan, to make the best use of all available information, and to engage relevant experts. -

Understanding the Space of Research
Biostatistics and Biometrics Open Access Journal ISSN: 2573-2633 Review Article Biostat Biometrics Open Acc J Volume 4 Issue 4 - January 2018 Copyright © All rights are reserved by Dhritikesh C DOI: 10.19080/BBOAJ.2018.04.555642 Understanding the Space of Research Dhritikesh C* Department of Statistics, Handique Girls’ College, India Submission: October 20, 2017; Published: January 22, 2018 *Corresponding author: Dhritikesh Chakrabarty, Department of Statistics, Handique Girls’ College, India Email: Abstract The theory of research, the history of the beginning of whose development was lost in the dust of antiquity, has been found to be the vital player in playing the role of developing knowledge everywhere. At the current stage of human civilization, research has become an unavoidable activity but also a partner/helper for most of the problems in the society. A journey has been made for understanding the space of research and essential component of each and every branch/field of academic world. The research is not only an unavoidable component of academic specifically on the meaning of research, definition of research, characteristics of research, methodology of research, types of research etc. This paperKeywords: is based Research; on some Research of the findings process; obtained Characteristics; in the study. Methodology; Types Meaning and Definition of Research Dictionary definitions o Research is a detailed study of a subject, especially in order to discover (new) information or reach a (new) o To search or investigate exhaustively. understanding. o Studious inquiry or examination; especially: o The word “research” is used to describe a number of investigation or experimentation aimed at the discovery and similar and often overlapping activities involving a search interpretation of facts, revision of accepted theories or laws for information.