Review Article CD4+T Cells: Differentiation and Functions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Interleukin 22 Pathway Interacts with Mutant KRAS to Promote Poor Prognosis in Colon Cancer
Author Manuscript Published OnlineFirst on May 19, 2020; DOI: 10.1158/1078-0432.CCR-19-1086 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. The interleukin 22 pathway interacts with mutant KRAS to promote poor prognosis in colon cancer Authors: Sarah McCuaig1, David Barras,2, Elizabeth Mann1, Matthias Friedrich1, Samuel Bullers1, Alina Janney1, Lucy C. Garner1, Enric Domingo3, Viktor Hendrik Koelzer3,4,5, Mauro Delorenzi2,6,7, Sabine Tejpar8, Timothy Maughan9, Nathaniel R. West1, Fiona Powrie1 Affiliations: 1 Kennedy Institute of Rheumatology, University of Oxford, Oxford UK. 2 SIB Swiss Institute of Bioinformatics, Bioinformatics Core Facility, Lausanne, Switzerland. 3Department of Oncology, University of Oxford, Oxford UK. 4Nuffield Department of Medicine, University of Oxford, Oxford UK. 5Department of Pathology and Molecular Pathology, University and University Hospital Zurich, Zurich Switzerland. 6 Ludwig Center for Cancer Research, University of Lausanne, Lausanne, Switzerland. 7 Department of Oncology, Faculty of Biology and Medicine, University of Lausanne, Lausanne Switzerland. 8 Molecular Digestive Oncology, KU Leuven, Belgium. 9 CRUK/MRC Oxford Institute for Radiation Oncology, University of Oxford, Oxford, UK. Downloaded from clincancerres.aacrjournals.org on September 26, 2021. © 2020 American Association for Cancer Research. Author Manuscript Published OnlineFirst on May 19, 2020; DOI: 10.1158/1078-0432.CCR-19-1086 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Correspondence to: Professor Fiona Powrie; Kennedy Institute of Rheumatology, University of Oxford, Roosevelt Drive, Headington, Oxford, OX3 7YF, UK. Email: [email protected] Conflicts of Interest: S.M., N.R.W., and F.P. -

IL-25-Induced Activities IL-17RB and IL-17RA in Mediating Identification of Functional Roles for Both
Identification of Functional Roles for Both IL-17RB and IL-17RA in Mediating IL-25-Induced Activities This information is current as Erika A. Rickel, Lori A. Siegel, Bo-Rin Park Yoon, James B. of September 29, 2021. Rottman, David G. Kugler, David A. Swart, Penny M. Anders, Joel E. Tocker, Michael R. Comeau and Alison L. Budelsky J Immunol 2008; 181:4299-4310; ; doi: 10.4049/jimmunol.181.6.4299 Downloaded from http://www.jimmunol.org/content/181/6/4299 References This article cites 30 articles, 16 of which you can access for free at: http://www.jimmunol.org/content/181/6/4299.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 29, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2008 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Identification of Functional Roles for Both IL-17RB and IL-17RA in Mediating IL-25-Induced Activities Erika A. -

The Interleukin 22 Pathway Interacts with Mutant KRAS to Promote Poor Prognosis in Colon Cancer
Author Manuscript Published OnlineFirst on May 19, 2020; DOI: 10.1158/1078-0432.CCR-19-1086 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. The interleukin 22 pathway interacts with mutant KRAS to promote poor prognosis in colon cancer Authors: Sarah McCuaig1, David Barras,2, Elizabeth Mann1, Matthias Friedrich1, Samuel Bullers1, Alina Janney1, Lucy C. Garner1, Enric Domingo3, Viktor Hendrik Koelzer3,4,5, Mauro Delorenzi2,6,7, Sabine Tejpar8, Timothy Maughan9, Nathaniel R. West1, Fiona Powrie1 Affiliations: 1 Kennedy Institute of Rheumatology, University of Oxford, Oxford UK. 2 SIB Swiss Institute of Bioinformatics, Bioinformatics Core Facility, Lausanne, Switzerland. 3Department of Oncology, University of Oxford, Oxford UK. 4Nuffield Department of Medicine, University of Oxford, Oxford UK. 5Department of Pathology and Molecular Pathology, University and University Hospital Zurich, Zurich Switzerland. 6 Ludwig Center for Cancer Research, University of Lausanne, Lausanne, Switzerland. 7 Department of Oncology, Faculty of Biology and Medicine, University of Lausanne, Lausanne Switzerland. 8 Molecular Digestive Oncology, KU Leuven, Belgium. 9 CRUK/MRC Oxford Institute for Radiation Oncology, University of Oxford, Oxford, UK. Downloaded from clincancerres.aacrjournals.org on September 30, 2021. © 2020 American Association for Cancer Research. Author Manuscript Published OnlineFirst on May 19, 2020; DOI: 10.1158/1078-0432.CCR-19-1086 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Correspondence to: Professor Fiona Powrie; Kennedy Institute of Rheumatology, University of Oxford, Roosevelt Drive, Headington, Oxford, OX3 7YF, UK. Email: [email protected] Conflicts of Interest: S.M., N.R.W., and F.P. -

ABD-Derived Protein Blockers of Human IL-17 Receptor a As Non-Igg Alternatives for Modulation of IL-17-Dependent Pro-Inflammatory Axis
International Journal of Molecular Sciences Article ABD-Derived Protein Blockers of Human IL-17 Receptor A as Non-IgG Alternatives for Modulation of IL-17-Dependent Pro-Inflammatory Axis Marie Hlavniˇcková 1, Milan Kuchaˇr 1, Radim Osiˇcka 2, Lucie Vaˇnková 1, Hana Petroková 1, Michal Malý 1, Jiˇrí Cernˇ ý 3 , Petr Arenberger 4 and Petr Malý 1,* 1 Laboratory of Ligand Engineering, Institute of Biotechnology of the Czech Academy of Sciences, v. v. i., BIOCEV Research Center, Pru˚myslová 595, 252 50 Vestec, Czech Republic; [email protected] (M.H.); [email protected] (M.K.); [email protected] (L.V.); [email protected] (H.P.); [email protected] (M.M.) 2 Laboratory of Molecular Biology of the Bacterial Pathogens, Institute of Microbiology, Czech Academy of Sciences, v. v. i., Vídeˇnská 1083, 142 20 Prague, Czech Republic; [email protected] 3 Laboratory of Structural Bioinformatics of Proteins, Institute of Biotechnology of the Czech Academy of Sciences, v. v. i., BIOCEV Research Center, Pru˚myslová 595, 252 50 Vestec, Czech Republic; [email protected] 4 Department of Dermatology and Venereology, Faculty Hospital of Královské Vinohrady, Šrobárova 50, 100 34 Prague, Czech Republic; [email protected] * Correspondence: [email protected]; Tel.: +420-325-873-763 Received: 10 September 2018; Accepted: 6 October 2018; Published: 9 October 2018 Abstract: Interleukin 17 (IL-17) and its cognate receptor A (IL-17RA) play a crucial role in Th17 cells-mediated pro-inflammatory pathway and pathogenesis of several autoimmune disorders including psoriasis. -

IL-17RA-Signaling Modulates CD8+ T Cell Survival and Exhaustion During 2 Trypanosoma Cruzi Infection
bioRxiv preprint doi: https://doi.org/10.1101/314336; this version posted May 11, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. 1 IL-17RA-signaling modulates CD8+ T cell survival and exhaustion during 2 Trypanosoma cruzi infection 3 Jimena Tosello Boari1,2, Cintia L. Araujo Furlan1,2, Facundo Fiocca Vernengo1,2, Constanza 4 Rodriguez1,2, María C. Ramello1,2, María C. Amezcua Vesely1,2, Melisa Gorosito Serrán1,2, 5 Nicolás G. Nuñez3,4, Wilfrid Richer3,4, Eliane Piaggio3,4, Carolina L. Montes1,2, Adriana 6 Gruppi1,2, Eva V. Acosta Rodríguez1,2*. 7 1 Departamento de Bioquímica Clínica. Facultad de Ciencias Químicas, Universidad 8 Nacional de Córdoba, Córdoba, X5000HUA. Argentina. 9 2 Centro de Investigaciones en Bioquímica Clínica e Inmunología. CONICET. Córdoba, 10 X5000HUA. Argentina. 11 3 SiRIC TransImm «Translational Immunotherapy Team», Translational Research 12 Department, Research Center, PSL Research University, INSERM U932, Institut Curie, 13 Paris, 75005. France. 14 4 Centre d’Investigation Clinique Biothérapie CICBT 1428, Institut Curie, Paris, 75005. 15 France. 16 Running title: IL-17RA signaling regulates CD8+ T cell responses to T. cruzi 17 *Corresponding autor: 18 Eva V. Acosta Rodríguez 19 [email protected] 20 1 bioRxiv preprint doi: https://doi.org/10.1101/314336; this version posted May 11, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Il-17 Receptor Composition 1995)
RESEARCH HIGHLIGHTS Nature Reviews Immunology | Published online 23 November 2015; doi:10.1038/nri.2015.2 shown to signal through IL-17RA, of the IL-17 family, whereas IL-17RC Journal club which was also unrelated to other seems to be specific for IL-17 and known cytokine receptors (Yao et al., IL-17F. Nonetheless, there are few IL-17 RECEPTOR COMPOSITION 1995). However, it was unclear how phenotypic differences between mice IL-17RA operated at a molecular level. lacking IL-17RA or IL-17RC, and It has now been a decade since the Toy et al. approached this issue by patients with null mutations in either paradigm-shifting discovery of T helper reconstituting fibroblasts from Il17ra–/– subunit also have remarkably similar 17 (TH17) cells in 2005 forced everyone mice with human IL-17RA. They defects, with their major phenotype working on effector CD4+ T cells to observed that IL-17-induced signalling being chronic mucosal candidiasis reinterpret their findings in the context could only be restored in these (Puel et al., 2011; Ling et al., 2015). of this interleukin-17 (IL-17)-producing fibroblasts upon co-expression of Thus, this important study by Toy et al. TH cell population. We now know that human IL-17RC with human IL-17RA, clarified the constituents of the IL-17 IL-17-induced signalling has a crucial but not upon co-expression of other receptor complex, which set the stage role in immunity to extracellular IL-17R family members. As the Il17ra–/– for a deeper understanding of its microorganisms, particularly to fungal cells expressed endogenous mouse biological activities. -
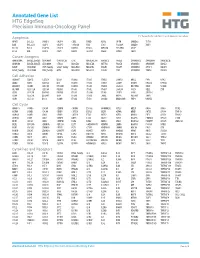
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel For Research Use Only. Not for use in diagnostic procedures. Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 -

Interleukin-17C Promotes Th17 Cell Responses and Autoimmune Disease Via Interleukin-17 Receptor E
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Immunity Article Interleukin-17C Promotes Th17 Cell Responses and Autoimmune Disease via Interleukin-17 Receptor E Seon Hee Chang,1,2 Joseph M. Reynolds,1,2 Bhanu P. Pappu,1 Guangjie Chen,1 Gustavo J. Martinez,1 and Chen Dong1,* 1Department of Immunology and Center for Inflammation and Cancer, MD Anderson Cancer Center, Houston, TX 77054-1901, USA 2These authors contributed equally to this work *Correspondence: [email protected] DOI 10.1016/j.immuni.2011.09.010 SUMMARY IL-23 (Langrish et al., 2005) for their differentiation and cytokine expression. Although several interleukin-17 (IL-17) family Th17 cells, via production of IL-17A and IL-17F, promote the members and their receptors have been recently development of autoimmune diseases while protecting the appreciated as important regulators in inflammatory host against bacterial and fungal infections (Kolls and Linde´ n, diseases, the function of other IL-17 cytokines and 2004). IL-17A and IL-17F signal through the IL-17 receptor A IL-17 receptor-like molecules is unclear. Here we (IL-17RA)-IL-17RC complex (Hu et al., 2010; Toy et al., 2006). show that an IL-17 cytokine family member, IL-17C, Although IL-17RA is expressed ubiquitously, IL-17RC expres- sion is dominant in nonhematopoietic cells (Kuestner et al., was induced in a Th17 cell-dependent autoimmune 2007). IL-17A and IL-17F target epithelial cells or fibroblasts to disease and was required for its pathogenesis. produce arrays of cytokines and chemokines including IL-6 IL-17C bound to IL-17RE, a member of IL-17 receptor and CXCL1 (Chang and Dong, 2009; Gaffen, 2008). -

Epigenetic Changes in Human Model KMT2A Leukemias Highlight Early Events During Leukemogenesis
Epigenetic changes in human model KMT2A leukemias highlight early events during leukemogenesis Thomas Milan1, Magalie Celton1, Karine Lagacé1, Élodie Roques1, Safia Safa-Tahar-Henni1, Eva Bresson2,3,4, Anne Bergeron2,3,4, Josée Hebert5,6, Soheil Meshinchi7, Sonia Cellot8, Frédéric Barabé2,3,4, 1,6 Brian T Wilhelm Author contributions: BTW and TM designed the study, analyzed data and drafted the manuscript. TM and KL performed ChIP-seq and ATAC-seq, analysed the data and generated figures. MC performed Methyl-seq experiments, analysed data, generated figures and drafted the manuscript. SS-T-H assisted with the ATAC-seq analysis and generation of figures. KL, ER, EB, and AB helped with molecular biology work, cloning and cell culture. EB, AB and FB generated the model on mice from CD34+ cells. SM provided expression and clinical data for patient samples and JH and SC collected, characterized, and banked the local patient samples used in this study. Affiliations: 1Laboratory for High Throughput Biology, Institute for Research in Immunology and Cancer, Montréal, QC, Canada 2Centre de recherche en infectiologie du CHUL, Centre de recherche du CHU de Québec – Université Laval, Québec City, QC, Canada, 3CHU de Québec – Université Laval – Hôpital Enfant-Jésus; Québec City, QC, Canada; 4Department of Medicine, Université Laval, Quebec City, QC, Canada 5Division of Hematology-Oncology and Leukemia Cell Bank of Quebec, Maisonneuve-Rosemont Hospital, Montréal, QC, Canada, 6Department of Medicine, Université de Montréal, Montréal, QC, Canada, 7Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA 8Department of pediatrics, division of Hematology, Ste-Justine Hospital, Montréal, QC, Canada Running head: Early epigenetic changes in KM3-AML *Correspondence to: Brian T Wilhelm Institute for Research in Immunology and Cancer Université de Montréal P.O. -

Copy Number Variation of IL17RA Gene And
Aghaei et al. BMC Medical Genetics (2020) 21:147 https://doi.org/10.1186/s12881-020-01078-y RESEARCH ARTICLE Open Access Copy number variation of IL17RA gene and its association with the ankylosing spondylitis risk in Iranian patients: a case- control study Hamideh Aghaei1,2, Elham Farhadi2,3, Maryam Akhtari2, Sara Shahba2, Shayan Mostafaei4, Ahmadreza Jamshidi2, Shiva Poursani2, Mahdi Mahmoudi2,3* and Mohammad Hossein Nicknam1,5* Abstract Background: Ankylosing spondylitis (AS) is considered as a subtype of spondyloarthritis (SpA) that mainly leads to fatigue, stiffness, spinal ankylosis, and impaired physical functions with reduced quality of life. Interleukin (IL)-17A provokes additional inflammatory mediators and recruits immune cells to the inflamed site. IL17 expression increased in various inflammatory disorders including psoriasis, rheumatoid arthritis, multiple sclerosis, crohn’s disease, and ankylosing spondylitis. The current study aimed to evaluate the association of IL17RA copy number changes with the susceptibility to AS and their correlation to IL17RA expression in Iranian population. Methods: IL17RA copy number genotyping assessments were carried out in 455 AS patients and 450 healthy controls, using custom TaqMan CNV assays. TaqMan primers and probe were located in Chr.22:17109553 based on pre-designed IL17RA Copy Number Assay ID, Hs02339506_cn. mRNA expression of IL17RA was also measured by SYBR Green real-time polymerase chain reaction (PCR). Results: A IL17RA copy number loss (< 2) was associated with AS compared to 2 copies as reference (OR:2.18, 95% CI: (1.38–3.44), P-value < 0.001) and increased the risk of AS. IL17RA mRNA expression showed a significant increase in peripheral blood mononuclear cells (PBMCs) of all AS individuals than controls. -

T Cell Receptor Signaling Pathway and Cytokine-Cytokine Receptor Interaction Affect the Rehabilitation Process After Respiratory Syncytial Virus Infection
T cell receptor signaling pathway and cytokine-cytokine receptor interaction affect the rehabilitation process after respiratory syncytial virus infection Zuanhao Qian*, Zhenglei Zhang* and Yingying Wang Department of Pediatrics, Taikang Xianlin Drum Tower Hospital, Nanjing, China * These authors contributed equally to this work. ABSTRACT Background. Respiratory syncytial virus (RSV) is the main cause of respiratory tract infection, which seriously threatens the health and life of children. This study is conducted to reveal the rehabilitation mechanisms of RSV infection. Methods. E-MTAB-5195 dataset was downloaded from EBI ArrayExpress database, including 39 acute phase samples in the acute phase of infection and 21 samples in the recovery period. Using the limma package, differentially expressed RNAs (DE- RNAs) were analyzed. The significant modules were identified using WGCNA package, and the mRNAs in them were conducted with enrichment analysis using DAVID tool. Afterwards, co-expression network for the RNAs involved in the significant modules was built by Cytoscape software. Additionally, RSV-correlated pathways were searched from Comparative Toxicogenomics Database, and then the pathway network was constructed. Results. There were 2,489 DE-RNAs between the two groups, including 2,386 DE- mRNAs and 103 DE-lncRNAs. The RNAs in the black, salmon, blue, tan and turquoise modules correlated with stage were taken as RNA set1. Meanwhile, the RNAs in brown, blue, magenta and pink modules related to disease severity were defined as RNA set2. In the pathway networks, CD40LG and RASGRP1 co-expressed Submitted 14 January 2019 with LINC00891/LINC00526/LINC01215 were involved in the T cell receptor sig- Accepted 6 May 2019 Published 12 June 2019 naling pathway, and IL1B, IL1R2, IL18, and IL18R1 co-expressed with BAIAP2- AS1/CRNDE/LINC01503/SMIM25 were implicated in cytokine-cytokine receptor Corresponding author Zuanhao Qian, interaction. -

Molecular Mechanisms of Synergy Between IL-13 and IL-17A In
Molecular Mechanisms of Synergy Between IL-13 and IL-17A in Severe Asthma A dissertation submitted to the Graduate School of the University of Cincinnati in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Immunology Graduate Program of the College of Medicine by Sara L. Hall M.S. University of Cincinnati 2011 Committee Chair: Ian P. Lewkowich, Ph.D. 1. Abstract Increased IL-17A production has been associated with more severe asthma; however, the mechanisms whereby IL-17A can contribute to IL-13-driven pathology in asthmatic patients remain unclear. In this thesis, we sought to elucidate the molecular mechanisms by which IL- 17A enhances IL-13-dependent airway pathology in patients with severe asthma using in vivo and in vitro systems. We have found that compared to mice given intratracheal (i.t.) IL-13 alone, those co-exposed to IL-13 + IL-17A demonstrate enhanced airway hyperresponsiveness (AHR), mucus production, airway inflammation, and IL-13-induced gene expression. In vitro, IL-17A directly enhanced IL-13-induced gene expression in asthma-relevant murine and human cells. In contrast to the exacerbating effect of IL-17A on IL-13-driven responses, co-treatment with IL-13 diminished IL-17A-driven gene expression in vivo and in vitro. Mechanistically, in vivo and in primary human and murine cells, the IL-17A mediated increase in IL-13-induced gene expression was associated with a rapid increase in IL-13-driven signal transducer and activator of transcription (STAT)6 phosphorylation. Disrupting protein-tyrosine phosphatase function using Na3VO4 abrogated IL-17A- mediated enhancement of IL-13-driven STAT6 phosphorylation, suggesting that the ability of IL-17A to augment IL-13 activity was driven by changes in protein-tyrosine phosphatase activity.