Parallel Processing Here at the School of Statistics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Different Unix Contexts
The different Unix contexts • User-level • Kernel “top half” - System call, page fault handler, kernel-only process, etc. • Software interrupt • Device interrupt • Timer interrupt (hardclock) • Context switch code Transitions between contexts • User ! top half: syscall, page fault • User/top half ! device/timer interrupt: hardware • Top half ! user/context switch: return • Top half ! context switch: sleep • Context switch ! user/top half Top/bottom half synchronization • Top half kernel procedures can mask interrupts int x = splhigh (); /* ... */ splx (x); • splhigh disables all interrupts, but also splnet, splbio, splsoftnet, . • Masking interrupts in hardware can be expensive - Optimistic implementation – set mask flag on splhigh, check interrupted flag on splx Kernel Synchronization • Need to relinquish CPU when waiting for events - Disk read, network packet arrival, pipe write, signal, etc. • int tsleep(void *ident, int priority, ...); - Switches to another process - ident is arbitrary pointer—e.g., buffer address - priority is priority at which to run when woken up - PCATCH, if ORed into priority, means wake up on signal - Returns 0 if awakened, or ERESTART/EINTR on signal • int wakeup(void *ident); - Awakens all processes sleeping on ident - Restores SPL a time they went to sleep (so fine to sleep at splhigh) Process scheduling • Goal: High throughput - Minimize context switches to avoid wasting CPU, TLB misses, cache misses, even page faults. • Goal: Low latency - People typing at editors want fast response - Network services can be latency-bound, not CPU-bound • BSD time quantum: 1=10 sec (since ∼1980) - Empirically longest tolerable latency - Computers now faster, but job queues also shorter Scheduling algorithms • Round-robin • Priority scheduling • Shortest process next (if you can estimate it) • Fair-Share Schedule (try to be fair at level of users, not processes) Multilevel feeedback queues (BSD) • Every runnable proc. -
Operating Systems
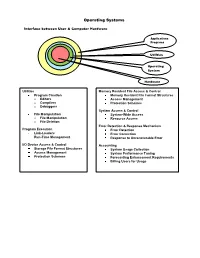
Operating Systems Interface between User & Computer Hardware Applications Programs Utilities Operating System Hardware Utilities Memory Resident File Access & Control Program Creation Memory Resident File Format Structures o Editors Access Management o Compilers Protection Schemes o Debuggers System Access & Control File Manipulation System-Wide Access o File Manipulation Resource Access o File Deletion Error Detection & Response Mechanism Program Execution Error Detection Link-Loaders Error Correction Run-Time Management Response to Unrecoverable Error I/O Device Access & Control Accounting Storage File Format Structures System Usage Collection Access Management System Performance Tuning Protection Schemes Forecasting Enhancement Requirements Billing Users for Usage Resource Manager O/S KernelKernel I/O Controller Printers, Keyboards, I/O Controller Monitors, Portions of Cameras, the O/S Etc. currently in use Computer Main System Memory Portions of Various I/O Application Devices Programs Currently in use Operating System Data Application Programs I/O Controller Storage Processor Processor Data Processor Processor Operation Allocation of Main Memory is made jointly by both the O/S and Memory Management Hardware O/S controls access to I/O devices by Application Programs O/S controls access to and use of files O/S controls access to and use of the processors, i.e., how much time can be allocated to the execution of a particular Application Program Classification of Operating Systems Interactive O/S Keyboard & Monitor Access to O/S Immediate, -

Backup and Restore Cisco Prime Collaboration Provisioning
Backup and Restore Cisco Prime Collaboration Provisioning This section explains the following: • Perform Backup and Restore, page 1 • Back Up the Single-Machine Provisioning Database, page 2 • Restore the Single-Machine Provisioning Database, page 3 • Schedule Backup Using the Provisioning User Interface, page 5 Perform Backup and Restore Cisco Prime Collaboration Provisioning allows you to backup your data and restore it. You can schedule periodic backups using the Provisioning UI (Schedule Backup Using the Provisioning User Interface, on page 5). There are two backup and restore scenarios; select the set of procedures that matches your scenario: • Backup and restore on a single machine, with the same installation or a new installation. For this scenario, see Schedule Backup Using the Provisioning User Interface, on page 5. Note When backing up files, you should place the files on a different file server. Also, you should burn the backup data onto a CD. Cisco Prime Collaboration Provisioning allows you to back up system data and restore it on a different system in the event of total system failure. To restore the backup from another system, the following prerequisites must be met: • Ensure that the server to which data is restored has the same MAC address as that of the system that was backed up (the IP address and the hostname can be different). • If you are unable to assign the MAC address of the original system (the one that was backed up) to another system, contact the Engineering Team for information on a new license file (for a new MAC address). Cisco Prime Collaboration Provisioning Install and Upgrade Guide, 12.3 1 Backup and Restore Cisco Prime Collaboration Provisioning Back Up the Single-Machine Provisioning Database • The procedure to backup and restore data on a different system is the same as the procedure to backup and restore data on the same system. -

FIFI-LS Highlights Robert Minchin FIFI-LS Highlights
FIFI-LS Highlights Robert Minchin FIFI-LS Highlights • Nice face-on galaxies where [CII] traces star formation [CII] traces star formation in M51 Pineda et al. 2018 [CII] in NGC 6946 Bigiel et al. 2020 [CII] particularly important as H2 & SFR tracer in inter-arm regions FIFI-LS Highlights • Nice face-on galaxies where [CII] traces star formation • Galaxies where [CII] doesn’t trace star formation [CII] from shocks & turbulence in NGC 4258 • [CII] seen along X-ray/radio ‘arms’ of NGC 4258, associated with past or present jet activity • [CII] excited by shocks and turbulence associated with the jet impacting the disk • NOT star formation Appleton et al. 2018 Excess [CII] in HE 1353-1917 1011 HE 1353-1917 1% MUSE RGB Images: 0.1 % • HE 1353-1917 1010 iband[OIII] Hα 0.02 % has a large 109 excess in HE 0433-1028 L[CII]/LFIR 8 10 excess 1σ 10 × 2σ /L 3C 326 3σ HE 1029-1831 • AGN ionization [CII] 7 10 HE 1108-2813 L cone intercepts HE 2211-3903 106 the cold SOFIA AGN Hosts galactic disk AGN Hosts High z 105 LINERs [Brisbin+15] Normal Galaxies LIRGs ULIRGs [Decarli+18] 104 108 109 1010 1011 1012 1013 1014 L /L Smirnova-Pinchukova et al. 2019 FIR Shock-excited [CII] in NGC 2445 [CII] in the ring of NGC 2445 is enhanced compared to PAHs, showing a shock excitement origin rather than star formation Fadda & Appleton, 2020, AAS Meeting 235 FIFI-LS Highlights • Nice face-on galaxies where [CII] traces star formation • Galaxies where [CII] doesn’t trace star formation • Galaxies with gas away from the plane Extraplanar [CII] in edge-on galaxies • Edge-on galaxies NGC 891 & NGC 5907 observed by Reach et al. -

Chapter 10 Introduction to Batch Files
Instructor’s Manual Chapter 10 Lecture Notes Introduction to Batch Files Chapter 10 Introduction to Batch Files LEARNING OBJECTIVES 1. Compare and contrast batch and interactive processing. 2. Explain how batch files work. 3. Explain the purpose and function of the REM, ECHO, and PAUSE commands. 4. Explain how to stop or interrupt the batch file process. 5. Explain the function and use of replaceable parameters in batch files. 6. Explain the function of pipes, filters, and redirection in batch files. STUDENT OUTCOMES 1. Use Edit to write batch files. 2. Use COPY CON to write batch files. 3. Write and execute a simple batch file. 4. Write a batch file to load an application program. 5. Use the REM, PAUSE, and ECHO commands in batch files. 6. Terminate a batch file while it is executing. 7. Write batch files using replaceable parameters. 8. Write a batch file using pipes, filters, and redirection. CHAPTER SUMMARY 1. Batch processing means running a series of instructions without interruption. 2. Interactive processing allows the user to interface directly with the computer and update records immediately. 3. Batch files allow a user to put together a string of commands and execute them with one command. 4. Batch files must have the .BAT or .CMD file extension. 5. Windows looks first internally for a command, then for a .COM files extension, then for a .EXE file extension, and finally for a .BAT or .CMD file extension. 6. Edit is a full-screen text editor used to write batch files. 7. A word processor, if it has a means to save files in ASCII, can be used to write batch files. -

Uni Hamburg – Mainframe Summit 2010 Z/OS – the Mainframe Operating System
Uni Hamburg – Mainframe Summit 2010 z/OS – The Mainframe Operating System Appendix 2 – JES and Batchprocessing Redelf Janßen IBM Technical Sales Mainframe Systems [email protected] © Copyright IBM Corporation 2010 Course materials may not be reproduced in whole or in part without the prior written permission of IBM. 4.0.1 Introduction to the new mainframe Chapter 7: Batch processing and the Job Entry Subsystem (JES) © Copyright IBM Corp., 2010. All rights reserved. Introduction to the new mainframe Chapter 7 objectives Be able to: • Give an overview of batch processing and how work is initiated and managed in the system. • Explain how the job entry subsystem (JES) governs the flow of work through a z/OS system. © Copyright IBM Corp., 2010. All rights reserved. 3 Introduction to the new mainframe Key terms in this chapter • batch processing • procedure • execution • purge • initiator • queue • job • spool • job entry subsystem (JES) • symbolic reference • output • workload manager (WLM) © Copyright IBM Corp., 2010. All rights reserved. 4 Introduction to the new mainframe What is batch processing? Much of the work running on z/OS consists of programs called batch jobs. Batch processing is used for programs that can be executed: • With minimal human interaction • At a scheduled time or on an as-needed basis. After a batch job is submitted to the system for execution, there is normally no further human interaction with the job until it is complete. © Copyright IBM Corp., 2010. All rights reserved. 5 Introduction to the new mainframe What is JES? In the z/OS operating system, JES manages the input and output job queues and data. -

Approaches to Optimize Batch Processing on Z/OS
Front cover Approaches to Optimize Batch Processing on z/OS Apply the latest z/OS features Analyze bottlenecks Evaluate critical path Alex Louwe Kooijmans Elsie Ramos Jan van Cappelle Lydia Duijvestijn Tomohiko Kaneki Martin Packer ibm.com/redbooks Redpaper International Technical Support Organization Approaches to Optimize Batch Processing on z/OS October 2012 REDP-4816-00 Note: Before using this information and the product it supports, read the information in “Notices” on page v. First Edition (October 2012) This edition applies to all supported z/OS versions and releases. This document created or updated on October 24, 2012. © Copyright International Business Machines Corporation 2012. All rights reserved. Note to U.S. Government Users Restricted Rights -- Use, duplication or disclosure restricted by GSA ADP Schedule Contract with IBM Corp. Contents Notices . .v Trademarks . vi Preface . vii The team who wrote this paper . vii Now you can become a published author, too! . viii Comments welcome. viii Stay connected to IBM Redbooks . ix Chapter 1. Getting started . 1 1.1 The initial business problem statement. 2 1.2 How to clarify the problem statement . 3 1.3 Process to formulate a good problem statement . 3 1.4 How to create a good business case . 5 1.5 Analysis methodology . 6 1.5.1 Initialization . 6 1.5.2 Analysis. 6 1.5.3 Implementation . 10 Chapter 2. Analysis steps. 13 2.1 Setting the technical strategy . 14 2.2 Understanding the batch landscape . 15 2.2.1 Identifying where batch runs and the available resources . 15 2.2.2 Job naming conventions . 15 2.2.3 Application level performance analysis. -

Sleep 2.1 Manual
Sleep 2.1 Manual "If you put a million monkeys at a million keyboards, one of them will eventually write a Java program. The rest of them will write Perl programs." -- Anonymous Raphael Mudge Sleep 2.1 Manual Revision: 06.02.08 Released under a Creative Commons Attribution-ShareAlike 3.0 License (see http://creativecommons.org/licenses/by-sa/3.0/us/) You are free: • to Share -- to copy, distribute, display, and perform the work • to Remix -- to make derivative works Under the following conditions: Attribution. You must attribute this work to Raphael Mudge with a link to http://sleep.dashnine.org/ Share Alike. If you alter, transform, or build upon this work, you may distribute the resulting work only under the same, similar or a compatible license. • For any reuse or distribution, you must make clear to others the license terms of this work. The best way to do this is with a link to the license. • Any of the above conditions can be waived if you get permission from the copyright holder. • Apart from the remix rights granted under this license, nothing in this license impairs or restricts the author's moral rights. Your fair use and other rights are in no way affected by the above. Table of Contents Introduction................................................................................................. 1 I. What is Sleep?...................................................................................................1 II. Manual Conventions......................................................................................2 III. -

Introduction to Unix Part I: the Essentials
Introduction to Unix Part I: The Essentials Frederick J Tan Bioinformatics Research Faculty Carnegie Institution of Washington, Department of Embryology 9 April 2013 Unix, Linux, Ubuntu, Oh My! 2 A Three Hour Tour Part I: The Essentials client-server model, command-line interface, navigation, viewing files, searching, finding help Part II: Special Topics account settings, multi-tasking, programming, installing programs, file systems, system administration 3 The Awesome Power of the Client-Server Model 4 A Tale of Two Interfaces Command Line Graphical Terminal.app, Unix shell Finder.app, Galaxy breadth, cutting-edge discovery, visualization 5 Running Programs Using the Command-Line Interface command-line graphical type in the name of the to run a program program and hit <ENTER> double click on an icon (e.g. bowtie2) type in options after the program, to modify how a click on check boxes, before hitting <ENTER> program operates select from pull-down menus (e.g. bowtie2 --very-sensitive) 6 The Anatomy of a Shell Prompt workshop@ubuntu:~$ The text in between the colon (:) The $ symbol indicates The symbol and the dollar sign ($) indicates that the server is ready to indicates where what what directory you are in. perform a command. you type in will appear. /home/workshop$ $ ssh [email protected] 7 Task 1: Connect to your server and start top with -i option Start VirtualBox Start Ubuntu Secure SHell $ ssh [email protected] <ENTER> <SPACE> Shell Prompt /home/workshop$ <TAB> Start top -i $ top -i <CTRL> <UP> <DOWN> 8 Task 2: Figure -
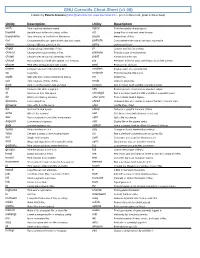
GNU Coreutils Cheat Sheet (V1.00) Created by Peteris Krumins ([email protected], -- Good Coders Code, Great Coders Reuse)
GNU Coreutils Cheat Sheet (v1.00) Created by Peteris Krumins ([email protected], www.catonmat.net -- good coders code, great coders reuse) Utility Description Utility Description arch Print machine hardware name nproc Print the number of processors base64 Base64 encode/decode strings or files od Dump files in octal and other formats basename Strip directory and suffix from file names paste Merge lines of files cat Concatenate files and print on the standard output pathchk Check whether file names are valid or portable chcon Change SELinux context of file pinky Lightweight finger chgrp Change group ownership of files pr Convert text files for printing chmod Change permission modes of files printenv Print all or part of environment chown Change user and group ownership of files printf Format and print data chroot Run command or shell with special root directory ptx Permuted index for GNU, with keywords in their context cksum Print CRC checksum and byte counts pwd Print current directory comm Compare two sorted files line by line readlink Display value of a symbolic link cp Copy files realpath Print the resolved file name csplit Split a file into context-determined pieces rm Delete files cut Remove parts of lines of files rmdir Remove directories date Print or set the system date and time runcon Run command with specified security context dd Convert a file while copying it seq Print sequence of numbers to standard output df Summarize free disk space setuidgid Run a command with the UID and GID of a specified user dir Briefly list directory -

Unix Quickref.Dvi
Summary of UNIX commands Table of Contents df [dirname] display free disk space. If dirname is omitted, 1. Directory and file commands 1994,1995,1996 Budi Rahardjo ([email protected]) display all available disks. The output maybe This is a summary of UNIX commands available 2. Print-related commands in blocks or in Kbytes. Use df -k in Solaris. on most UNIX systems. Depending on the config- uration, some of the commands may be unavailable 3. Miscellaneous commands du [dirname] on your site. These commands may be a commer- display disk usage. cial program, freeware or public domain program that 4. Process management must be installed separately, or probably just not in less filename your search path. Check your local documentation or 5. File archive and compression display filename one screenful. A pager similar manual pages for more details (e.g. man program- to (better than) more. 6. Text editors name). This reference card, obviously, cannot de- ls [dirname] scribe all UNIX commands in details, but instead I 7. Mail programs picked commands that are useful and interesting from list the content of directory dirname. Options: a user's point of view. 8. Usnet news -a display hidden files, -l display in long format 9. File transfer and remote access mkdir dirname Disclaimer make directory dirname The author makes no warranty of any kind, expressed 10. X window or implied, including the warranties of merchantabil- more filename 11. Graph, Plot, Image processing tools ity or fitness for a particular purpose, with regard to view file filename one screenfull at a time the use of commands contained in this reference card. -

MTS on Wikipedia Snapshot Taken 9 January 2011
MTS on Wikipedia Snapshot taken 9 January 2011 PDF generated using the open source mwlib toolkit. See http://code.pediapress.com/ for more information. PDF generated at: Sun, 09 Jan 2011 13:08:01 UTC Contents Articles Michigan Terminal System 1 MTS system architecture 17 IBM System/360 Model 67 40 MAD programming language 46 UBC PLUS 55 Micro DBMS 57 Bruce Arden 58 Bernard Galler 59 TSS/360 60 References Article Sources and Contributors 64 Image Sources, Licenses and Contributors 65 Article Licenses License 66 Michigan Terminal System 1 Michigan Terminal System The MTS welcome screen as seen through a 3270 terminal emulator. Company / developer University of Michigan and 7 other universities in the U.S., Canada, and the UK Programmed in various languages, mostly 360/370 Assembler Working state Historic Initial release 1967 Latest stable release 6.0 / 1988 (final) Available language(s) English Available programming Assembler, FORTRAN, PL/I, PLUS, ALGOL W, Pascal, C, LISP, SNOBOL4, COBOL, PL360, languages(s) MAD/I, GOM (Good Old Mad), APL, and many more Supported platforms IBM S/360-67, IBM S/370 and successors History of IBM mainframe operating systems On early mainframe computers: • GM OS & GM-NAA I/O 1955 • BESYS 1957 • UMES 1958 • SOS 1959 • IBSYS 1960 • CTSS 1961 On S/360 and successors: • BOS/360 1965 • TOS/360 1965 • TSS/360 1967 • MTS 1967 • ORVYL 1967 • MUSIC 1972 • MUSIC/SP 1985 • DOS/360 and successors 1966 • DOS/VS 1972 • DOS/VSE 1980s • VSE/SP late 1980s • VSE/ESA 1991 • z/VSE 2005 Michigan Terminal System 2 • OS/360 and successors