Two Novel Protein O-Glucosyltransferases That Modify Sites Distinct from POGLUT1 and Affect Notch Trafficking and Signaling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bacteria Belonging to Pseudomonas Typographi Sp. Nov. from the Bark Beetle Ips Typographus Have Genomic Potential to Aid in the Host Ecology
insects Article Bacteria Belonging to Pseudomonas typographi sp. nov. from the Bark Beetle Ips typographus Have Genomic Potential to Aid in the Host Ecology Ezequiel Peral-Aranega 1,2 , Zaki Saati-Santamaría 1,2 , Miroslav Kolaˇrik 3,4, Raúl Rivas 1,2,5 and Paula García-Fraile 1,2,4,5,* 1 Microbiology and Genetics Department, University of Salamanca, 37007 Salamanca, Spain; [email protected] (E.P.-A.); [email protected] (Z.S.-S.); [email protected] (R.R.) 2 Spanish-Portuguese Institute for Agricultural Research (CIALE), 37185 Salamanca, Spain 3 Department of Botany, Faculty of Science, Charles University, Benátská 2, 128 01 Prague, Czech Republic; [email protected] 4 Laboratory of Fungal Genetics and Metabolism, Institute of Microbiology of the Academy of Sciences of the Czech Republic, 142 20 Prague, Czech Republic 5 Associated Research Unit of Plant-Microorganism Interaction, University of Salamanca-IRNASA-CSIC, 37008 Salamanca, Spain * Correspondence: [email protected] Received: 4 July 2020; Accepted: 1 September 2020; Published: 3 September 2020 Simple Summary: European Bark Beetle (Ips typographus) is a pest that affects dead and weakened spruce trees. Under certain environmental conditions, it has massive outbreaks, resulting in attacks of healthy trees, becoming a forest pest. It has been proposed that the bark beetle’s microbiome plays a key role in the insect’s ecology, providing nutrients, inhibiting pathogens, and degrading tree defense compounds, among other probable traits. During a study of bacterial associates from I. typographus, we isolated three strains identified as Pseudomonas from different beetle life stages. In this work, we aimed to reveal the taxonomic status of these bacterial strains and to sequence and annotate their genomes to mine possible traits related to a role within the bark beetle holobiont. -

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Aberrant Sialylation in Cancer: Biomarker and Potential Target for Therapeutic Intervention?
cancers Review Aberrant Sialylation in Cancer: Biomarker and Potential Target for Therapeutic Intervention? Silvia Pietrobono * and Barbara Stecca * Tumor Cell Biology Unit, Core Research Laboratory, Institute for Cancer Research and Prevention (ISPRO), Viale Pieraccini 6, 50139 Florence, Italy * Correspondence: [email protected] (S.P.); [email protected] (B.S.); Tel.: +39-055-7944568 (S.P.); +39-055-7944567 (B.S.) Simple Summary: Sialylation is a post-translational modification that consists in the addition of sialic acid to growing glycan chains on glycoproteins and glycolipids. Aberrant sialylation is an established hallmark of several types of cancer, including breast, ovarian, pancreatic, prostate, colorectal and lung cancers, melanoma and hepatocellular carcinoma. Hypersialylation can be the effect of increased activity of sialyltransferases and results in an excess of negatively charged sialic acid on the surface of cancer cells. Sialic acid accumulation contributes to tumor progression by several paths, including stimulation of tumor invasion and migration, and enhancing immune evasion and tumor cell survival. In this review we explore the mechanisms by which sialyltransferases promote cancer progression. In addition, we provide insights into the possible use of sialyltransferases as biomarkers for cancer and summarize findings on the development of sialyltransferase inhibitors as potential anti-cancer treatments. Abstract: Sialylation is an integral part of cellular function, governing many biological processes Citation: Pietrobono, S.; Stecca, B. including cellular recognition, adhesion, molecular trafficking, signal transduction and endocytosis. Aberrant Sialylation in Cancer: Sialylation is controlled by the levels and the activities of sialyltransferases on glycoproteins and Biomarker and Potential Target for lipids. Altered gene expression of these enzymes in cancer yields to cancer-specific alterations of Therapeutic Intervention? Cancers glycoprotein sialylation. -

Insulin in Insulin-Secreti
OPEN Rab2A is a pivotal switch protein that SUBJECT AREAS: promotes either secretion or MEMBRANE TRAFFICKING ER-associated degradation of (pro)insulin ORGANELLES in insulin-secreting cells Received 1 1,2 1 8 July 2014 Taichi Sugawara , Fumi Kano & Masayuki Murata Accepted 14 October 2014 1Department of Life Sciences, Graduate School of Arts and Sciences, The University of Tokyo, Tokyo 153-8902, Japan, 2PRESTO, Japan Science and Technology Agency, Saitama 332-0012, Japan. Published 7 November 2014 Rab2A, a small GTPase localizing to the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC), regulates COPI-dependent vesicular transport from the ERGIC. Rab2A knockdown inhibited glucose-stimulated insulin secretion and concomitantly enlarged the ERGIC in insulin-secreting cells. Large Correspondence and aggregates of polyubiquitinated proinsulin accumulated in the cytoplasmic vicinity of a unique large requests for materials spheroidal ERGIC, designated the LUb-ERGIC. Well-known components of ER-associated degradation should be addressed to (ERAD) also accumulated at the LUb-ERGIC, creating a suitable site for ERAD-mediated protein quality M.M. (mmurata@bio. control. Moreover, chronically high glucose levels, which induced the enlargement of the LUb-ERGIC and c.u-tokyo.ac.jp) ubiquitinated protein aggregates, impaired Rab2A activity by promoting dissociation from its effector, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), in response to poly (ADP-ribosyl)ation of GAPDH. The inactivation of Rab2A relieved glucose-induced ER stress and inhibited ER stress-induced apoptosis. Collectively, these results suggest that Rab2A is a pivotal switch that controls whether insulin should be secreted or degraded at the LUb-ERGIC and Rab2A inactivation ensures alleviation of ER stress and cell survival under chronic glucotoxicity. -

Trehalose and Trehalose-Based Polymers for Environmentally Benign, Biocompatible and Bioactive Materials
Molecules 2008, 13, 1773-1816; DOI: 10.3390/molecules13081773 OPEN ACCESS molecules ISSN 1420-3049 www.mdpi.org/molecules Review Trehalose and Trehalose-based Polymers for Environmentally Benign, Biocompatible and Bioactive Materials Naozumi Teramoto 1, *, Navzer D. Sachinvala 2, † and Mitsuhiro Shibata 1 1 Department of Life and Environmental Sciences, Faculty of Engineering, Chiba Institute of Technology, 2-17-1 Tsudanuma, Narashino, Chiba 275-0016, Japan; E-mail: [email protected] 2 Retired, Southern Regional Research Center, USDA-ARS, New Orleans, LA, USA; Home: 2261 Brighton Place, Harvey, LA 70058; E-mail: [email protected] † Dedicated to Professor George Christensen, Department of Psychology, Winona State University, Winona, MN, USA. * Author to whom correspondence should be addressed; E-mail: [email protected]. Received: 13 July 2008 / Accepted: 11 August 2008 / Published: 21 August 2008 Abstract: Trehalose is a non-reducing disaccharide that is found in many organisms but not in mammals. This sugar plays important roles in cryptobiosis of selaginella mosses, tardigrades (water bears), and other animals which revive with water from a state of suspended animation induced by desiccation. The interesting properties of trehalose are due to its unique symmetrical low-energy structure, wherein two glucose units are bonded face-to-face by 1J1-glucoside links. The Hayashibara Co. Ltd., is credited for developing an inexpensive, environmentally benign and industrial-scale process for the enzymatic conversion of α-1,4-linked polyhexoses to α,α-D-trehalose, which made it easy to explore novel food, industrial, and medicinal uses for trehalose and its derivatives. -

Induced Structural Changes in a Multifunctional Sialyltransferase
Biochemistry 2006, 45, 2139-2148 2139 Cytidine 5′-Monophosphate (CMP)-Induced Structural Changes in a Multifunctional Sialyltransferase from Pasteurella multocida†,‡ Lisheng Ni,§ Mingchi Sun,§ Hai Yu,§ Harshal Chokhawala,§ Xi Chen,*,§ and Andrew J. Fisher*,§,| Department of Chemistry and the Section of Molecular and Cellular Biology, UniVersity of California, One Shields AVenue, DaVis, California 95616 ReceiVed NoVember 23, 2005; ReVised Manuscript ReceiVed December 19, 2005 ABSTRACT: Sialyltransferases catalyze reactions that transfer a sialic acid from CMP-sialic acid to an acceptor (a structure terminated with galactose, N-acetylgalactosamine, or sialic acid). They are key enzymes that catalyze the synthesis of sialic acid-containing oligosaccharides, polysaccharides, and glycoconjugates that play pivotal roles in many critical physiological and pathological processes. The structures of a truncated multifunctional Pasteurella multocida sialyltransferase (∆24PmST1), in the absence and presence of CMP, have been determined by X-ray crystallography at 1.65 and 2.0 Å resolutions, respectively. The ∆24PmST1 exists as a monomer in solution and in crystals. Different from the reported crystal structure of a bifunctional sialyltransferase CstII that has only one Rossmann domain, the overall structure of the ∆24PmST1 consists of two separate Rossmann nucleotide-binding domains. The ∆24PmST1 structure, thus, represents the first sialyltransferase structure that belongs to the glycosyltransferase-B (GT-B) structural group. Unlike all other known GT-B structures, however, there is no C-terminal extension that interacts with the N-terminal domain in the ∆24PmST1 structure. The CMP binding site is located in the deep cleft between the two Rossmann domains. Nevertheless, the CMP only forms interactions with residues in the C-terminal domain. -

Downloaded 18 July 2014 with a 1% False Discovery Rate (FDR)
UC Berkeley UC Berkeley Electronic Theses and Dissertations Title Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer Permalink https://escholarship.org/uc/item/0t47b9ws Author Spiciarich, David Publication Date 2017 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer by David Spiciarich A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Chemistry in the Graduate Division of the University of California, Berkeley Committee in charge: Professor Carolyn R. Bertozzi, Co-Chair Professor David E. Wemmer, Co-Chair Professor Matthew B. Francis Professor Amy E. Herr Fall 2017 Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer © 2017 by David Spiciarich Abstract Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer by David Spiciarich Doctor of Philosophy in Chemistry University of California, Berkeley Professor Carolyn R. Bertozzi, Co-Chair Professor David E. Wemmer, Co-Chair Changes in glycosylation have long been appreciated to be part of the cancer phenotype; sialylated glycans are found at elevated levels on many types of cancer and have been implicated in disease progression. However, the specific glycoproteins that contribute to cell surface sialylation are not well characterized, specifically in bona fide human cancer. Metabolic and bioorthogonal labeling methods have previously enabled enrichment and identification of sialoglycoproteins from cultured cells and model organisms. The goal of this work was to develop technologies that can be used for detecting changes in glycoproteins in clinical models of human cancer. -
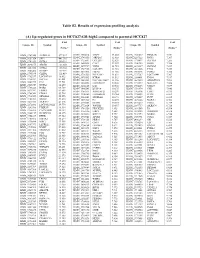
(A) Up-Regulated Genes in HCC827-GR-High2 Compared to Parental HCC827
Table S2. Results of expression profiling analysis (A) Up-regulated genes in HCC827-GR-high2 compared to parental HCC827 Fold Fold Fold Unique ID Symbol Unique ID Symbol Unique ID Symbol change* change* change* ILMN_1709348 ALDH1A1 577.587 ILMN_2310814 MAPT 13.003 ILMN_1741017 PIP4K2B 7.331 ILMN_1651354 SPP1 441.316 ILMN_1748650 MRPL45 12.988 ILMN_3237623 RNY1 7.297 ILMN_1701831 GSTA1 260.591 ILMN_1755897 UGT2B7 12.629 ILMN_1734897 SLC4A4 7.285 ILMN_1658835 CAV2 12.357 ILMN_1746359 RERG 7.280 ILMN_2094875 ABCB1 183.050 ILMN_1678939 VNN2 11.935 ILMN_1671337 SLC2A5 7.257 ILMN_3251540 GSTA2 145.982 ILMN_1729905 GAL3ST1 11.910 ILMN_1691606 LYG2 7.254 ILMN_2062468 IGFBP7 127.721 ILMN_1672536 FBLN1 11.716 ILMN_1785646 PMP22 7.246 ILMN_1795190 CLDN2 111.439 ILMN_1796339 PLEKHA2 11.631 ILMN_1737387 LOC728441 7.207 ILMN_1782937 LOC647169 98.612 ILMN_1676563 HTRA1 11.592 ILMN_1684401 FMO1 7.117 ILMN_1754247 SLC3A1 81.001 ILMN_3263423 LOC100129027 11.346 ILMN_1687035 ADAMTSL4 7.098 ILMN_1662795 CA2 79.581 ILMN_1694898 LOC653857 10.906 ILMN_2153572 MAGEA3 7.086 ILMN_2168747 GSTA2 66.250 ILMN_2404625 LAT 10.560 ILMN_1784283 USH1C 7.079 ILMN_1764228 DAB2 64.709 ILMN_1666546 DUSP14 10.375 ILMN_1731374 CPE 7.046 ILMN_1675797 EPDR1 63.605 ILMN_1764571 ARHGAP23 10.299 ILMN_1765446 EMP3 6.933 ILMN_1708341 PDZK1 59.714 ILMN_3200140 LOC645638 10.284 ILMN_1754002 IL1F8 6.863 ILMN_1713529 SEMA6A 52.575 ILMN_3244343 SNORA21 10.171 ILMN_1878007 FUT9 6.835 ILMN_1708391 NR1H4 43.218 ILMN_1671489 PC 10.075 ILMN_1699208 NAP1L1 6.763 ILMN_2412336 AKR1C2 42.826 ILMN_2404688 -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

(12) STANDARD PATENT (11) Application No. AU 2015215937 B2 (19) AUSTRALIAN PATENT OFFICE
(12) STANDARD PATENT (11) Application No. AU 2015215937 B2 (19) AUSTRALIAN PATENT OFFICE (54) Title Metabolically engineered organisms for the production of added value bio-products (51) International Patent Classification(s) C12N 15/52 (2006.01) C12P 19/26 (2006.01) C12P 19/18 (2006.01) C12P 19/30 (2006.01) (21) Application No: 2015215937 (22) Date of Filing: 2015.08.21 (43) Publication Date: 2015.09.10 (43) Publication Journal Date: 2015.09.10 (44) Accepted Journal Date: 2017.03.16 (62) Divisional of: 2011278315 (71) Applicant(s) Universiteit Gent (72) Inventor(s) MAERTENS, Jo;BEAUPREZ, Joeri;DE MEY, Marjan (74) Agent / Attorney Griffith Hack, GPO Box 3125, Brisbane, QLD, 4001, AU (56) Related Art Trinchera, M. et al., 'Dictyostelium cytosolic fucosyltransferase synthesizes H type 1 trisaccharide in vitro', FEBS Letters, 1996, Vol. 395, pages 68-72 GenBank accession no. AF279134, 6 May 2002 van der Wel, H. et al., 'A bifunctional diglycosyltransferase forms the Fuc#1,2Gal#1,3-disaccharide on Skpl in the cytoplasm of Dictyostelium', The Journal of Biological Chemistry, 2002, Vol. 277, No. 48, pages 46527-46534 WO 2010/070104 Al Abstract The present invention relates to genetically engineered organisms, especially microorganisms such as bacteria and yeasts, for the production of added value bio-products such as specialty saccharide, activated saccharide, nucleoside, glycoside, glycolipid or glycoprotein. More specifically, the present invention relates to host cells that are metabolically engineered so that they can produce said valuable specialty products in large quantities and at a high rate by bypassing classical technical problems that occur in biocatalytical or fermentative production processes. -

Synthesis of Fructooligosaccharides by Isla4, a Truncated Inulosucrase
Peña-Cardeña et al. BMC Biotechnology (2015) 15:2 DOI 10.1186/s12896-015-0116-1 RESEARCH ARTICLE Open Access Synthesis of Fructooligosaccharides by IslA4, a truncated inulosucrase from Leuconostoc citreum Arlen Peña-Cardeña, María Elena Rodríguez-Alegría, Clarita Olvera and Agustín López Munguía* Abstract Background: IslA4 is a truncated single domain protein derived from the inulosucrase IslA, which is a multidomain fructosyltransferase produced by Leuconostoc citreum. IslA4 can synthesize high molecular weight inulin from sucrose, with a residual sucrose hydrolytic activity. IslA4 has been reported to retain the product specificity of the multidomain enzyme. Results: Screening experiments to evaluate the influence of the reactions conditions, especially the sucrose and enzyme concentrations, on IslA4 product specificity revealed that high sucrose concentrations shifted the specificity of the reaction towards fructooligosaccharides (FOS) synthesis, which almost eliminated inulin synthesis and led to a considerable reduction in sucrose hydrolysis. Reactions with low IslA4 activity and a high sucrose activity allowed for high levels of FOS synthesis, where 70% sucrose was used for transfer reactions, with 65% corresponding to transfructosylation for the synthesis of FOS. Conclusions: Domain truncation together with the selection of the appropriate reaction conditions resulted in the synthesis of various FOS, which were produced as the main transferase products of inulosucrase (IslA4). These results therefore demonstrate that bacterial fructosyltransferase