Immunolocation and Enzyme Activity Analysis of Cryptosporidium Parvum
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Why the –Omic Future of Apicomplexa Should Include Gregarines Julie Boisard, Isabelle Florent
Why the –omic future of Apicomplexa should include Gregarines Julie Boisard, Isabelle Florent To cite this version: Julie Boisard, Isabelle Florent. Why the –omic future of Apicomplexa should include Gregarines. Biology of the Cell, Wiley, 2020, 10.1111/boc.202000006. hal-02553206 HAL Id: hal-02553206 https://hal.archives-ouvertes.fr/hal-02553206 Submitted on 24 Apr 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Article title: Why the –omic future of Apicomplexa should include Gregarines. Names of authors: Julie BOISARD1,2 and Isabelle FLORENT1 Authors affiliations: 1. Molécules de Communication et Adaptation des Microorganismes (MCAM, UMR 7245), Département Adaptations du Vivant (AVIV), Muséum National d’Histoire Naturelle, CNRS, CP52, 57 rue Cuvier 75231 Paris Cedex 05, France. 2. Structure et instabilité des génomes (STRING UMR 7196 CNRS / INSERM U1154), Département Adaptations du vivant (AVIV), Muséum National d'Histoire Naturelle, CP 26, 57 rue Cuvier 75231 Paris Cedex 05, France. Short Title: Gregarines –omics for Apicomplexa studies -

Studies in Cryptosporidium
STUDIES IN CRYPTOSPORIDIUM: MAINTENANCE OF STABLE POPULATIONS THROUGH IN VIVO PROPOGATION AND MOLECULAR DETECTION STRATEGIES DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of the Ohio State University By Norma E. Ramirez, M.P.H. ! ! ! ! The Ohio State University 2005 Dissertation Committee: Approved by Dr. Srinand Sreevatsan, Adviser Dr. Y.M. Saif ______________________________ Dr. Roger W. Stich Adviser Dr. Lucy A. Ward Graduate Program in Veterinary Preventive Medicine ABSTRACT Cryptosporidiosis, an infection caused by several genotypically and phenotypically diverse Cryptosporidium species, is a serious enteric disease of animals and humans worldwide. The current understanding of cryptosporidiosis, transmission, diagnosis, treatment and prevention measures for this disease is discussed. Contaminated water represents the major source of Cryptosporidium infections for humans. Manure from cattle can be a major source of Cryptosporidium oocysts. Oocysts transport to surface water can occur through direct fecal contamination, surface transport from land-applied manure or leaching through the soil to groundwater. Identification of Cryptosporidium species and genotypes facilitates determining the origin of the oocysts and to recognize sources of infection in outbreak situations and the risk factors associated with transmission. Very few studies have applied isolation methods to field samples because of difficulties with detection of oocysts in environmental samples. The objective of this study was to develop an easy method that can be applied to field samples to rapidly detect the presence of Cryptosporidium oocysts and identify their species. A molecular detection system that included an oocyst recovery method combined with spin column DNA extraction, followed by PCR- hybridization for detection and a Real-Time PCR-melting curve analysis for species ii assignment. -

Diversity of Extracellular Proteins During the Transition from the 'Proto
1 Diversity of extracellular proteins during the transition from the ‘proto-apicomplexan’ alveolates to the apicomplexan obligate parasites THOMAS J. TEMPLETON1,2* and ARNAB PAIN3,4 1 Department of Protozoology, Institute of Tropical Medicine (NEKKEN), Nagasaki University, 1-12-4 Sakamoto, Nagasaki 852-8523, Japan 2 Department of Microbiology and Immunology, Weill Cornell Medical College, New York 10021, USA 3 Pathogen Genomics Laboratory, Biological and Environmental Sciences and Engineering (BESE) Division, King Abdullah University of Science and Technology (KAUST), Thuwal, Jeddah 23955-6900, Kingdom of Saudi Arabia 4 Global Station for Zoonosis Control, Global Institution for Collaborative Research and Education (GI-CoRE), Hokkaido University, N20 W10 Kita-ku, Sapporo 001-0020, Japan (Received 22 May 2015; revised 12 August 2015; accepted 14 August 2015) SUMMARY The recent completion of high-coverage draft genome sequences for several alveolate protozoans – namely, the chromer- ids, Chromera velia and Vitrella brassicaformis; the perkinsid Perkinsus marinus; the apicomplexan, Gregarina niphandrodes, as well as high coverage transcriptome sequence information for several colpodellids, allows for new genome-scale com- parisons across a rich landscape of apicomplexans and other alveolates. Genome annotations can now be used to help in- terpret fine ultrastructure and cell biology, and guide new studies to describe a variety of alveolate life strategies, such as symbiosis or free living, predation, and obligate intracellular parasitism, as well to provide foundations to dissect the evo- lutionary transitions between these niches. This review focuses on the attempt to identify extracellular proteins which might mediate the physical interface of cell–cell interactions within the above life strategies, aided by annotation of the repertoires of predicted surface and secreted proteins encoded within alveolate genomes. -

Cryptosporidiosis: Biology, Pathogenesis and Disease Saul Tzipori A,*, Honorine Ward B
Microbes and Infection 4 (2002) 1047–1058 www.elsevier.com/locate/micinf Current Focus Cryptosporidiosis: biology, pathogenesis and disease Saul Tzipori a,*, Honorine Ward b a Division of Infectious Diseases, Tufts University School of Veterinary Medicine, North Grafton, MA 01536, USA b Division of Geographic Medicine/Infectious Diseases, New England Medical Center, Tufts University School of Medicine, Boston, MA 02111, USA Abstract Ninety-five years after discovery and after more than two decades of intense investigations, cryptosporidiosis, in many ways, remains enigmatic. Cryptosporidium infects all four classes of vertebrates and most likely all mammalian species. The speciation of the genus continues to be a challenge to taxonomists, compounded by many factors, including current technical difficulties and the apparent lack of host specificity by most, but not all, isolates and species. © 2002 Éditions scientifiques et médicales Elsevier SAS. All rights reserved. Keywords: Cryptosporidiosis; Cryptosporidium parvum; Diarrhea; Enteric infection; Opportunistic infection 1. Historical perspective occupies within the host cell membrane are the most obvious [6]. Although Cryptosporidium was first described in the Between 1980 and 1993, three broad entities of laboratory mouse by Tyzzer in 1907 [1], the medical and cryptosporidiosis became recognized [7]. The first was the veterinary significance of this protozoan was not fully revelation in 1980 that Cryptosporidium was, in fact, a appreciated for another 70 years. The interest in Cryptospo- common, yet serious, primary cause of outbreaks as well as ridium escalated tremendously over the last two decades, as sporadic cases of diarrhea in certain mammals [6]. From reflected in the number of publications, which increased 1983 onwards, with the onset of the AIDS epidemic, from 80 in 1983 to 2850 currently listed in MEDLINE. -

Minireview: Clinical Cryptosporidiosis
Experimental Parasitology 124 (2010) 138–146 Contents lists available at ScienceDirect Experimental Parasitology journal homepage: www.elsevier.com/locate/yexpr Minireview: Clinical cryptosporidiosis Rachel M. Chalmers a,*, Angharad P. Davies b a Head of UK Cryptosporidium Reference Unit, NPHS Microbiology Swansea, Singleton Hospital, Sketty Lane, Swansea SA2 8QA, UK b Clinical Senior Lecturer/Honorary Consultant Microbiologist, The School of Medicine, Swansea University, Swansea SA2 8PP, UK article info abstract Article history: Cryptosporidium has emerged as an important cause of diarrhoeal illness worldwide, especially amongst Received 24 October 2008 young children and patients with immune deficiencies. Usually presenting as a gastro-enteritis-like syn- Received in revised form 22 January 2009 drome, disease ranges in seriousness from mild to severe and signs and symptoms depend on the site of Accepted 5 February 2009 infection, nutritional and immune status of the host, and parasite-related factors. Sources and routes of Available online 11 February 2009 transmission are multiple, involving both zoonotic and anthroponotic spread, and facilitated by the resis- tance of the parasite to many commonly used disinfectants. Prevention and control measures are impor- Keywords: tant for the protection of vulnerable groups since treatment options are limited. This review covers the Apicomplexa life cycle, pathogenesis, clinical presentations, diagnosis, prevention and management of cryptosporidi- Cryptosporidium Cryptosporidiosis osis in humans. Humans Crown Copyright Ó 2009 Published by Elsevier Inc. All rights reserved. Cryptosporidum hominis Cryptosporidium parvum Genotypes Clinical features 1. Introduction varies geographically, and infection with unusual species and genotypes occurs in both immune-competent and immune-com- Cryptosporidiosis is the clinical disease, usually presenting as a promised populations (Cama et al., 2008). -

Cryptosporidium Muris, a Rodent Pathogen, Recovered from A
DISPATCHES recovered during the summer of 2002 in stools of an HIV- Cryptosporidium positive Peruvian woman with severe diarrhea. This find- ing was confirmed by light microscopy, polymerase chain muris, a Rodent reaction (PCR)–restriction fragment length polymorphism Pathogen, (RFLP), and DNA sequencing. The Study Recovered from a In 2002, we conducted a year-long collaborative study on the epidemiology of Cyclospora cayetanensis infec- Human in Perú tions in Perú. As part of that study, we collected approxi- mately 100 stool samples in 2.5% potassium dichromate Carol J. Palmer,* Lihua Xiao,† solution from persons in Lima and Iquitos with Angélica Terashima,‡ Humberto Guerra,‡ Cyclospora infection. Fecal samples were initially identi- Eduardo Gotuzzo,‡ Gustavo Saldías,§ fied as Cyclospora-positive in Lima, and then transported J. Alfredo Bonilla,* Ling Zhou,† Alan Lindquist,¶ to the United States for additional confirmation using wet and Steve J. Upton# mount and Nomarski interference contrast microscopy. Cryptosporidium muris, predominantly a rodent Two stool samples, which were taken on two sequential species of Cryptosporidium, is not normally considered a days from an HIV-positive woman who was 31 years of human pathogen. Recently, isolated human infections have age, contained oocysts that appeared, based on morpholo- been reported from Indonesia, Thailand, France, and gy, to be Cryptosporidium muris. Low numbers of Kenya. We report the first case of C. muris in a human in Cyclospora cayetanensis and Blastocystis hominis oocysts the Western Hemisphere. This species may be an emerg- were also identified in the stool samples. The ing zoonotic pathogen capable of infecting humans. Cryptosporidium muris infection was initially identified by using wet mount microscopy with oocysts (n=25) aver- ryptosporidiosis can be a debilitating diarrheal dis- aging 6.1 (± 0.3) x 8.4 (± 0.3) µM (range 5.6–6.4 x Cease. -
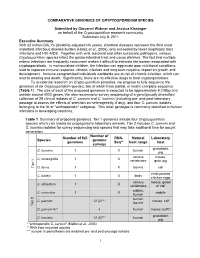
1 Comparative Genomics of Cryptosporidium Species
COMPARATIVE GENOMICS OF CRYPTOSPORIDIUM SPECIES Submitted by Giovanni Widmer and Jessica Kissinger on behalf of the Cryptosporidium research community Submitted July 8, 2011 Executive Summary With 62 million DALYs (disability-adjusted life years), diarrheal diseases represent the third most important infectious disease burden (Hotez et al., 2006), only exceeded by lower respiratory tract infections and HIV-AIDS. Together with viral, bacterial and other eukaryotic pathogens, various Cryptosporidium species infect the gastro-intestinal tract and cause diarrhea. The fact that multiple enteric infections are frequently concurrent makes it difficult to estimate the burden associated with cryptosporidiosis. In malnourished children, the infection can aggravate poor nutritional conditions, lead to impaired immune response, chronic infection and long-term negative impact on growth and development. Immune-compromised individuals worldwide are at risk of chronic infection, which can lead to wasting and death. Significantly, there are no effective drugs to treat cryptosporidiosis. To accelerate research on Cryptosporidium parasites, we propose to fully sequence the genomes of six Cryptosporidium species, two of which have partial, or nearly complete sequence (Table 1). The size of each of the proposed genomes is expected to be approximately 9.2 Mbp and contain around 4000 genes. We also recommend survey sequencing of a genotypically diversified collection of 28 clinical isolates of C. parvum and C. hominis (including pre- and post-laboratory passage to assess the effects of selection on heterogeneity, if any), and four C. parvum isolates belonging to the IIc or "anthroponotic" subgroup. This latter genotype is commonly identified in human infections in developing countries. Table 1 Summary of proposed genomes. -

Bovine Gastric and Intestinal Cryptosporidiosis: Present Situation
Bovine Gastric and Intestinal Cryptosporidiosis: Present Situation (Q) n 0 "'O Bruce C. Anderson '-< ......'"i University of Idaho, College ofAgriculture (JQ Department ofAnimal and Veterinary Science g > Caine Veterinary Teaching and Research Center 8 1020 East Homedale Road, Caldwell, ID 83605 (D ......'"i (") Introduction § >00 00 This writer intends to provide information which will by the fecal-oral route. The excreted oocyst survives well in 0 (") be useful or may be useful, next week in the practices of cool moist environments but does not need a sporulation ...... a...... cattle practitioners. Comments on epidemiology, control period outside the host; the oocysts are excreted fully spo 0 ~ and zoonotic potential of Cryptosporidium parvum (intes rulated and are immediately infective for susceptible hosts. 0 tinal pathogen) were solicited for an oral presentation at Significant is the fact that thin-walled oocysts within the 1-i; to the recent meeting of the American Association of Bovine host are auto-infective, thus providing a mechanism for 0 < Practitioners. But, from day-to-day bovine practice stand massive magnification of the infection within the host. In 5· point, there isn't much to say about this intestinal patho calves, the clinical diarrheal episode usually begins in the (D gen, despite the publication of about 1000 articles on ~ second week of life and usually is over by the end of week 3 ~ (") cryptosporidiosis in the past 10 years. In fact, William Cur of age. Oocyst shedding at a rate of about a million per ,-+-...... rent and Lynn Garcia stated recently in a review article1 ,-+-...... gram of feces occurs during the first several days of diar 0 ~ (357 references) that, "Despite the large number of recent rhea, and may continue at a lesser rate through week 4 (D '"i papers and the large number of laboratories throughout after birth. -

Cyclospora Cayetanensis—Major Outbreaks from Ready to Eat Fresh Fruits and Vegetables
foods Review Cyclospora Cayetanensis—Major Outbreaks from Ready to Eat Fresh Fruits and Vegetables Agni Hadjilouka 1,2 and Dimitris Tsaltas 2,* 1 EMBIO Diagnostics LTD., Athalassas 8b, 2018 Nicosia, Cyprus; [email protected] 2 Department of Agricultural Sciences, Biotechnology and Food Science, Cyprus University of Technology, Archbishop Kyprianos 30, 3036 Limassol, Cyprus * Correspondence: [email protected]; Tel.: +357-2500-2545 Received: 6 October 2020; Accepted: 18 November 2020; Published: 20 November 2020 Abstract: Cyclospora cayetanensis is a coccidian protozoan that causes cyclosporiasis, a severe gastroenteric disease, especially for immunocompromised patients, children, and the elderly. The parasite is considered as an emerging organism and a major contributor of gastroenteritis worldwide. Although the global prevalence of cyclosporiasis morbidity and mortality has not been assessed, global concern has arisen since diarrheal illness and gastroenteritis significantly affect both developing countries and industrialized nations. In the last two decades, an increasing number of foodborne outbreaks has been associated with the consumption of fresh produce that is difficult to clean thoroughly and is consumed without processing. Investigations of these outbreaks have revealed the necessity to increase the awareness in clinicians of this infection, since this protozoan is often ignored by surveillance systems, and to establish control measures to reduce contamination of fresh produce. In this review, the major cyclosporiasis outbreaks linked to the consumption of ready to eat fresh fruits and vegetables are presented. Keywords: Cyclospora cayetanensis; major outbreaks; fresh produce 1. Introduction Diarrhea is one of the leading causes of mortality worldwide. In 2016, it was responsible for the death of more than 1.6 million people, with 90% of the deaths being reported in South Asia and sub-Saharan Africa [1]. -

Occurrence and Genetic Diversity of Protist Parasites in Captive Non
animals Article Occurrence and Genetic Diversity of Protist Parasites in Captive Non-Human Primates, Zookeepers, and Free-Living Sympatric Rats in the Córdoba Zoo Conservation Centre, Southern Spain Pamela C. Köster 1, Alejandro Dashti 1 , Begoña Bailo 1, Aly S. Muadica 1,2 , Jenny G. Maloney 3, Mónica Santín 3, Carmen Chicharro 1 , Silvia Migueláñez 1, Francisco J. Nieto 1, David Cano-Terriza 4 , Ignacio García-Bocanegra 4 , Rafael Guerra 5, Francisco Ponce-Gordo 6, Rafael Calero-Bernal 7, David González-Barrio 1,7,* and David Carmena 1,* 1 Parasitology Reference and Research Laboratory, Spanish National Centre for Microbiology, 28220 Madrid, Spain; [email protected] (P.C.K.); [email protected] (A.D.); [email protected] (B.B.); [email protected] (A.S.M.); [email protected] (C.C.); [email protected] (S.M.); [email protected] (F.J.N.) 2 Departamento de Ciências e Tecnologia, Universidade Licungo, Quelimane 106, Zambézia, Mozambique 3 Environmental Microbial and Food Safety Laboratory, Agricultural Research Service, United States Department of Agriculture, Beltsville, MD 20705-2350, USA; [email protected] (J.G.M.); [email protected] (M.S.) 4 Animal Health and Zoonosis Research Group (GISAZ), Department of Animal Health, University of Córdoba, 14071 Córdoba, Spain; [email protected] (D.C.-T.); [email protected] (I.G.-B.) 5 Citation: Köster, P.C.; Dashti, A.; Veterinary Services, Córdoba Zoo Conservation Centre, 14071 Córdoba, Spain; Bailo, B.; Muadica, A.S.; Maloney, J.G.; [email protected] 6 Santín, M.; Chicharro, C.; Department of Microbiology and Parasitology, Faculty of Pharmacy, Complutense University of Madrid, 28040 Madrid, Spain; [email protected] Migueláñez, S.; Nieto, F.J.; 7 SALUVET, Department of Animal Health, Faculty of Veterinary, Complutense University of Madrid, Cano-Terriza, D.; et al. -

Cryptosporidium and Giardia in Africa: Current and Future Challenges Sylvia Afriyie Squire1,2 and Una Ryan1*
Squire and Ryan Parasites & Vectors (2017) 10:195 DOI 10.1186/s13071-017-2111-y REVIEW Open Access Cryptosporidium and Giardia in Africa: current and future challenges Sylvia Afriyie Squire1,2 and Una Ryan1* Abstract Cryptosporidium and Giardia are important causes of diarrhoeal illness. Adequate knowledge of the molecular diversity and geographical distribution of these parasites and the environmental and climatic variables that influence their prevalence is important for effective control of infection in at-risk populations, yet relatively little is known about the epidemiology of these parasites in Africa. Cryptosporidium is associated with moderate to severe diarrhoea and increased mortality in African countries and both parasites negatively affect child growth and development. Malnutrition and HIV status are also important contributors to the prevalence of Cryptosporidium and Giardia in African countries. Molecular typing of both parasites in humans, domestic animals and wildlife to date indicates a complex picture of both anthroponotic, zoonotic and spill-back transmission cycles that requires further investigation. For Cryptosporidium, the only available drug (nitazoxanide) is ineffective in HIV and malnourished individuals and therefore more effective drugs are a high priority. Several classes of drugs with good efficacy exist for Giardia, but dosing regimens are suboptimal and emerging resistance threatens clinical utility. Climate change and population growth are also predicted to increase both malnutrition and the prevalence of these parasites in water sources. Dedicated and co-ordinated commitments from African governments involving “One Health” initiatives with multidisciplinary teams of veterinarians, medical workers, relevant government authorities, and public health specialists working together are essential to control and prevent the burden of disease caused by these parasites. -

New Developments in Cryptosporidium Research
International Journal for Parasitology 45 (2015) 367–373 Contents lists available at ScienceDirect International Journal for Parasitology journal homepage: www.elsevier.com/locate/ijpara Invited Review New developments in Cryptosporidium research ⇑ Una Ryan a, ,1, Nawal Hijjawi b a School of Veterinary and Life Sciences, Vector- and Water-Borne Pathogen Research Group, Murdoch University, Murdoch, Western Australia 6150, Australia b Department of Medical Laboratory Sciences, Faculty of Allied Health Sciences, The Hashemite University PO Box 150459, Zarqa 13115, Jordan article info abstract Article history: Cryptosporidium is an enteric parasite that is considered the second greatest cause of diarrhoea and death Received 30 October 2014 in children after rotavirus. Currently, 27 species are recognised as valid and of these, Cryptosporidium Received in revised form 20 January 2015 hominis and Cryptosporidium parvum are responsible for the majority of infections in humans. Accepted 21 January 2015 Molecular and biological studies indicate that Cryptosporidium is more closely related to gregarine para- Available online 10 March 2015 sites rather than to coccidians. The identification of gregarine-like gamont stages and the ability of Cryptosporidium to complete its life cycle in the absence of host cells further confirm its relationship with Keywords: gregarines. This opens new avenues into the investigation of pathogenesis, epidemiology, treatment and Cryptosporidium control of Cryptosporidium. Effective drug treatments and vaccines are not yet available due, in part, to the Taxonomy Cell culture technical challenges of working on Cryptosporidium in the laboratory. Whole genome sequencing and Vaccines metabolomics have expanded our understanding of the biochemical requirements of this organism and Genomics have identified new drug targets.