Causes and Consequences of Genetic Robustness and Fragility in HIV-1 Proteins Suzannah J
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Developing a Single-Cycle Infectious System to Study an ERV-K Retroviral Envelope
Developing a Single-Cycle Infectious System to Study an ERV-K Retroviral Envelope Author: Rana Elias Akleh Persistent link: http://hdl.handle.net/2345/bc-ir:107695 This work is posted on eScholarship@BC, Boston College University Libraries. Boston College Electronic Thesis or Dissertation, 2017 Copyright is held by the author. This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License (http:// creativecommons.org/licenses/by-nc-sa/4.0). Developing a Single-Cycle Infectious System to Study an ERV-K Retroviral Envelope Rana Elias Akleh A thesis submitted to the faculty of the department of Biology in partial fulfillment of the requirements for the degree of Master of Science Boston College Morrissey College of Arts and Sciences Graduate School September 2017 © 2017 Rana Elias Akleh Developing a Single-Cycle Infectious System to Study an ERV-K Retroviral Envelope Rana Elias Akleh Advisor: Welkin Johnson, Ph.D. Endogenous Retroviruses (ERVs) are “fossilized” retroviruses of a once exogenous retrovirus located in the genome of extant vertebrates. Retroviral infection results in a provirus integration into the host genome. An infection of a germline cell could lead to the provirus potentially being inherited by the offspring of the infected individual. Once in the genome, the provirus becomes subject to evolutionary processes and can become either lost or fixed in a population, remaining as “fossils” long after the exogenous retrovirus has gone extinct23. Notably, 8% of the human genome consists of ERVs30. Human Endogenous Retrovirus Type K (HERV-K)(HML-2) family is of particular interest. HERV-K integrations are as old as 30-35 million years, endogenizing before the separation of humans and Old World Monkeys. -

Researchers Unveil New Monkey Model for HIV 2 March 2009
Researchers unveil new monkey model for HIV 2 March 2009 By altering just one gene in HIV-1, scientists have evolving genes, APOBEC3 and TRIM5, which succeeded in infecting pig-tailed macaque produce unusual classes of defensive proteins with monkeys with a human version of the virus that distinctive capabilities to fight retroviruses such as has until now been impossible to study directly in HIV. These genes, shared by humans and their animals. The new strain of HIV has already been simian forebears, have evolved mutations specific used to demonstrate one method for preventing to each species' unique history of retroviral battles. infection and, with a little tweaking, could be a In most simians, the APOBEC3 and TRIM5 valuable model for vetting vaccine candidates. proteins actually kill HIV on sight, making it impossible for researchers to study the virus in an A team of researchers led by Paul Bieniasz and animal model. Instead, they have studied HIV's Theodora Hatziioannou at The Rockefeller cousin, simian immunodeficiency virus (SIV), which University showed that two pig-tailed macaques, causes an AIDS-like disease in certain monkey given a common antiretroviral treatment one week species. But SIV shares only about half of its amino before and one week after being exposed to the acid sequence with HIV, making it a very imperfect newly engineered HIV, had no signs of infection. substitute for testing anti-HIV drugs and vaccines. "We're not saying we can save the world with Several labs have engineered hybrids called SHIVs antiretroviral pills. But this model will allow us to — SIVs that contain pieces of HIV DNA — but these start studying the best way to administer have problematic differences, too. -
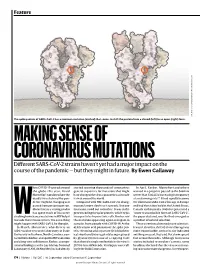
MAKING SENSE of CORONAVIRUS MUTATIONS Different SARS-Cov-2 Strains Haven’T Yet Had a Major Impact on the Course of the Pandemic — but They Might in Future
Feature SOURCE: STRUCTURAL DATA FROM K. SHEN & J. LUBAN K. SHEN FROM DATA STRUCTURAL SOURCE: The spike protein of SARS-CoV-2 has a common mutation (circled) that seems to shift the protein from a closed (left) to an open (right) form. MAKING SENSE OF CORONAVIRUS MUTATIONS Different SARS-CoV-2 strains haven’t yet had a major impact on the course of the pandemic — but they might in future. By Ewen Callaway hen COVID-19 spread around started scouring thousands of coronavirus In April, Korber, Montefiori and others the globe this year, David genetic sequences for mutations that might warned in a preprint posted to the bioRxiv Montefiori wondered how the have changed the virus’s properties as it made server that “D614G is increasing in frequency deadly virus behind the pan- its way around the world. at an alarming rate”1. It had rapidly become demic might be changing as it Compared with HIV, SARS-CoV-2 is chang- the dominant SARS-CoV-2 lineage in Europe passed from person to person. ing much more slowly as it spreads. But one and had then taken hold in the United States, Montefiori is a virologist who mutation stood out to Korber. It was in the Canada and Australia. D614G represented a has spent much of his career gene encoding the spike protein, which helps “more transmissible form of SARS-CoV-2”, Wstudying how chance mutations in HIV help it virus particles to penetrate cells. Korber saw the paper declared, one that had emerged as to evade the immune system. -
Croi 2021 Program Committee
General Information CONTENTS WELCOME . 2 General Information General Information OVERVIEW . 2 CONTINUING MEDICAL EDUCATION . 3 CONFERENCE SUPPORT . 4 VIRTUAL PLATFORM . 5 ON-DEMAND CONTENT AND WEBCASTS . 5 CONFERENCE SCHEDULE AT A GLANCE . 6 PRECONFERENCE SESSIONS . 9 LIVE PLENARY, ORAL, AND INTERACTIVE SESSIONS, AND ON-DEMAND SYMPOSIA BY DAY . 11 SCIENCE SPOTLIGHTS™ . 47 SCIENCE SPOTLIGHT™ SESSIONS BY CATEGORY . 109 CROI FOUNDATION . 112 IAS–USA . 112 CROI 2021 PROGRAM COMMITTEE . 113 Scientific Program Committee . 113 Community Liaison Subcommittee . 113 Former Members . 113 EXTERNAL REVIEWERS . .114 SCHOLARSHIP AWARDEES . 114 AFFILIATED OR PROXIMATE ACTIVITIES . 114 EMBARGO POLICIES AND SOCIAL MEDIA . 115 CONFERENCE ETIQUETTE . 115 ABSTRACT PROCESS Scientific Categories . 116 Abstract Content . 117 Presenter Responsibilities . 117 Abstract Review Process . 117 Statistics for Abstracts . 117 Abstracts Related to SARS-CoV-2 and Special Study Populations . 117. INDEX OF SPECIAL STUDY POPULATIONS . 118 INDEX OF PRESENTING AUTHORS . .122 . Version 9 .0 | Last Update on March 8, 2021 Printed in the United States of America . © Copyright 2021 CROI Foundation/IAS–USA . All rights reserved . ISBN #978-1-7320053-4-1 vCROI 2021 1 General Information WELCOME TO vCROI 2021 Welcome to vCROI 2021! The COVID-19 pandemic has changed the world for all of us in so many ways . Over the past year, we have had to put some of our HIV research on hold, learned to do our research in different ways using different tools, to communicate with each other in virtual formats, and to apply the many lessons in HIV research, care, and community advocacy to addressing the COVID-19 pandemic . Scientists and community stakeholders who have long been engaged in the endeavor to end the epidemic of HIV have pivoted to support and inform the unprecedented progress made in battle against SARS-CoV-2 . -

Final Scientific Programme
Final Scientific Programme Day 1 – Thursday, April 29 Arrival of conferees, registration, get-together party Session 1 Welcoming and Opening Remarks Chairman: Jan Svoboda 04:30 - 05:00 pm Jan Svoboda, Institute of Molecular Genetics, Prague Background and perspectives David Baltimore, California Institute of Technology, Pasadena 05:00 - 06:00 pm Keynote lecture 06:00 pm Welcome reception Day 2 – Friday, April 30 Session 2 Action at the Cell Surface Chairmen: John A.T. Young, Benjamin K. Chen 08:30 - 09:00 am John A.T. Young, Salk Institute, La Jolla Cellular factor involvement in retroviral replication 09:00 - 09:30 am Benjamin K. Chen, Mount Sinai School of Medicine, New York Spread of HIV through T cell virological synapses 09:30 - 09:45 am Paul Jolicoeur, Clinical Research Institute of Montreal Pathogenesis of HIV-1 Nef in thymocytes 09:45 - 10:00 am Michaela Rumlová, Institute of Organic Chemistry and Biochemistry, Prague Analysis of N-terminus of NC protein of M-PMV: its role in the particle assembly 10:00 - 10:30 am Coffee break Session 3 Identification of Essential Host Cell Factors Chairmen: Stephen P. Goff, Alan N. Engelmam 10:30 - 11:00 am Stephen P. Goff, Columbia University, New York HIV-1 Interactions with host proteins 11:00 - 11:30 am Alan N. Engelmam, Dana-Farber Cancer Institute, Boston HIV-1 preintegration complex function and host cell factors 11:30 - 11:45 am Peter Cherepanov, Imperial College London Structural basis for retroviral pre-integration complex assembly and strand transfer inhibitor action 11:45 - 12:00 am C. -

Molecular Ecology and Natural History of Simian Foamy Virus Infection in Wild-Living Chimpanzees
Molecular Ecology and Natural History of Simian Foamy Virus Infection in Wild-Living Chimpanzees Weimin Liu1, Michael Worobey2, Yingying Li1, Brandon F. Keele1, Frederic Bibollet-Ruche1, Yuanyuan Guo1, Paul A. Goepfert1, Mario L. Santiago3, Jean-Bosco N. Ndjango4, Cecile Neel5,6, Stephen L. Clifford7, Crickette Sanz8, Shadrack Kamenya9, Michael L. Wilson10, Anne E. Pusey11, Nicole Gross-Camp12, Christophe Boesch8, Vince Smith13, Koichiro Zamma14, Michael A. Huffman15, John C. Mitani16, David P. Watts17, Martine Peeters5, George M. Shaw1, William M. Switzer18, Paul M. Sharp19, Beatrice H. Hahn1* 1 Departments of Medicine and Microbiology, University of Alabama at Birmingham, Birmingham, Alabama, United States of America, 2 University of Arizona, Tucson, Arizona, United States of America, 3 Gladstone Institute for Virology and Immunology, University of California at San Francisco, San Francisco, California, United States of America, 4 Faculties of Sciences, University of Kisangani, Democratic Republic of Congo, 5 Institut de Recherche pour le De´veloppement (IRD) and University of Montpellier 1, Montpellier, France, 6 Projet Prevention du Sida ou Cameroun (PRESICA), Yaounde´, Cameroun, 7 Centre International de Recherches Medicales de Franceville (CIRMF), Franceville, Gabon, 8 Max-Planck Institute for Evolutionary Anthropology, Leipzig, Germany, 9 Gombe Stream Research Centre, The Jane Goodall Institute, Tanzania, 10 Department of Anthropology, University of Minnesota, Minneapolis, Minnesota, United States of America, 11 Jane Goodall Institute’s -

MAKING SENSE of CORONAVIRUS MUTATIONS Different SARS-Cov-2 Strains Haven’T Yet Had a Major Impact on the Course of the Pandemic — but They Might in Future
Feature SOURCE: STRUCTURAL DATA FROM K. SHEN & J. LUBAN K. SHEN FROM DATA STRUCTURAL SOURCE: The spike protein of SARS-CoV-2 has a common mutation (circled) that seems to shift the protein from a closed (left) to an open (right) form. MAKING SENSE OF CORONAVIRUS MUTATIONS Different SARS-CoV-2 strains haven’t yet had a major impact on the course of the pandemic — but they might in future. By Ewen Callaway hen COVID-19 spread around had already started scouring thousands of In April, Korber, Montefiori and others the globe this year, David coronavirus genetic sequences for mutations warned in a preprint posted to the bioRxiv Montefiori wondered how the that might have changed the virus’s properties server that “D614G is increasing in frequency deadly virus behind the pan- as it made its way around the world. at an alarming rate”1. It had rapidly become demic might be changing as it Compared with HIV, SARS-CoV-2 is chang- the dominant SARS-CoV-2 lineage in Europe passed from person to person. ing much more slowly as it spreads. But one and had then taken hold in the United States, Montefiori is a virologist who mutation stood out to Korber. It was in the Canada and Australia. D614G represented a has spent much of his career gene encoding the spike protein, which helps “more transmissible form of SARS-CoV-2”, Wstudying how chance mutations in HIV help it virus particles to penetrate cells. Korber saw the paper declared, one that had emerged as to evade the immune system. -

Assembly Models for Viral Capsids Based on Tiling Theory
Viruses under the mathematical microscope: MathematicalAn opportunity to demonstrate Virology the impact of mathematics in biology in the classroom environment in the classroom Reidun Twarock Departments of Mathematics and Biology York Cross-disciplinary Centre for Systems Analysis H. C. Ørsted Institute, Copenhagen, November 2018 Central Questions 1. How can mathematics help to make discoveries in virology and find novel anti-viral solutions? 2. Which aspects can be covered in the classroom? -> Suggestions are given in the Teacher’s Packs The biological challenge Viruses are responsible for a wide spectrum of devastating diseases in humans animals and plants. Examples: •HIV •Hepatitis C •Cancer-causing viruses •Picornaviruses linked with type 1 diabetes •Common Cold • Options for anti-viral interventions are limited. • Therapy resistant mutant strains provide a challenge for therapy Protein Containers Viral capsids are like Trojan horses, hiding the genome from the defense mechanisms of their host. = Challenges: • What are the mathematical rules underpinning their structure? • Can this insight be used to combat viruses by preventing their formation? Viruses and Geometry An understanding of Viral Geometry enables discovery in virology and creates new opportunities in bionanotechnology and anti-viral therapy Symmetry in Virology What is icosahedral symmetry? The icosahedron has •6 axes of 5-fold symmetry •10 axes of 3-fold symmetry •15 axes of 2-fold symmetry Part I: What are the mathematical rules? The architecture of larger viruses Caspar -

Molecular Ecology and Natural History of Simian Foamy Virus Infection in Wild-Living Chimpanzees
Molecular Ecology and Natural History of Simian Foamy Virus Infection in Wild-Living Chimpanzees Weimin Liu1, Michael Worobey2, Yingying Li1, Brandon F. Keele1, Frederic Bibollet-Ruche1, Yuanyuan Guo1, Paul A. Goepfert1, Mario L. Santiago3, Jean-Bosco N. Ndjango4, Cecile Neel5,6, Stephen L. Clifford7, Crickette Sanz8, Shadrack Kamenya9, Michael L. Wilson10, Anne E. Pusey11, Nicole Gross-Camp12, Christophe Boesch8, Vince Smith13, Koichiro Zamma14, Michael A. Huffman15, John C. Mitani16, David P. Watts17, Martine Peeters5, George M. Shaw1, William M. Switzer18, Paul M. Sharp19, Beatrice H. Hahn1* 1 Departments of Medicine and Microbiology, University of Alabama at Birmingham, Birmingham, Alabama, United States of America, 2 University of Arizona, Tucson, Arizona, United States of America, 3 Gladstone Institute for Virology and Immunology, University of California at San Francisco, San Francisco, California, United States of America, 4 Faculties of Sciences, University of Kisangani, Democratic Republic of Congo, 5 Institut de Recherche pour le De´veloppement (IRD) and University of Montpellier 1, Montpellier, France, 6 Projet Prevention du Sida ou Cameroun (PRESICA), Yaounde´, Cameroun, 7 Centre International de Recherches Medicales de Franceville (CIRMF), Franceville, Gabon, 8 Max-Planck Institute for Evolutionary Anthropology, Leipzig, Germany, 9 Gombe Stream Research Centre, The Jane Goodall Institute, Tanzania, 10 Department of Anthropology, University of Minnesota, Minneapolis, Minnesota, United States of America, 11 Jane Goodall Institute’s -

Paul D. Bieniasz, Ph.D
PAUL D. BIENIASZ, PH.D. INTRINSIC CELLULAR DEFENSES AGAINST RETROVIRAL ATTACK MARCH 9, 2017 4:00 P.M. 208 LIGHT HALL SPONSORED BY: DEPARTMENT OF PATHOLOGY, MICROBIOLOGY, & IMMUNOLOGY Upcoming Discovery Lecture: AJIT VARKI, MBBS (M.D.) University of California, San Diego Distinguished Professor of Medicine and Cellular & Molecular Medicine March 23, 2017 208 Light Hall / 4:00 P.M. 1190-4260-Institution-Discovery Lecture Series-Bieniasz-BK-CH.indd 1 2/8/17 12:25 PM PAUL D. BIENIASZ, PH.D. INVESTIGATOR, HHMI INTRINSIC CELLULAR DEFENSES PROFESSOR AND HEAD, LABORATORY OF RETROVIROLOGY, AGAINST RETROVIRAL ATTACK AARON DIAMOND AIDS RESEARCH CENTER, THE ROCKEFELLER UNIVERSITY Throughout their evolution, most eukaryotic organisms have Paul Bieniasz studies the molecular biology of retroviruses and frequently been colonized by retroviruses. Indeed, selection their interaction with host cells. His work on HIV-1 particle pressures imposed by ancient retroviral infections are likely assembly includes the initial discovery and characterization of responsible for shaping the array of host defense mechanisms that ESCRT pathway in HIV-1 budding, and the development of currently influence susceptibility to modern retroviruses such as imaging techniques to quantitatively describe HIV-1 particle HIV-1. The Bieniasz laboratory works on several types of intrinsic genesis. Most recently his group has redefined how HIV-1 selects its defenses to understand the mechanistic details by which they RNA genome for packaging into virion particles. The Bieniasz lab inhibit retrovirus replication, including host molecules that inhibit was among the first to demonstrate the existence of antiretroviral virion release, block nuclear import, and deplete viral RNA. factors (so called restriction factors) that target the capsid of incoming lentiviruses. -

DARWIN's Surprise
ANNALS OF SCIENCE Darwin’S SuRpRISE Why are evolutionary biologists bringing back extinct deadly viruses? by michael specter hierry Heidmann’s office, adjacent manity than viral diseases: yellow fever, composed of broken and disabled retro- to the laboratory he runs at the In- measles, and smallpox have been caus- viruses, which, millions of years ago, stitutT Gustave Roussy, on the southern ing epidemics for thousands of years. At managed to embed themselves in the edge of Paris, could pass for a museum of the end of the First World War, fifty DNA of our ancestors. They are called genetic catastrophe. Files devoted to the million people died of the Spanish flu; endogenous retroviruses, because once world’s most horrifying infectious dis- smallpox may have killed half a billion they infect the DNA of a species they eases fill the cabinets and line the shelves. during the twentieth century alone. become part of that species. One by one, There are thick folders for smallpox, Those viruses were highly infectious, though, after molecular battles that Ebola virus, and various forms of in- yet their impact was limited by their fe- raged for thousands of generations, they fluenza. sars is accounted for, as are rocity: a virus may destroy an entire cul- have been defeated by evolution. Like more obscure pathogens, such as feline ture, but if we die it dies, too. As a re- dinosaur bones, these viral fragments are leukemia virus, Mason-Pfizer monkey sult, not even smallpox possessed the fossils. Instead of having been buried in virus, and simian foamy virus, which is evolutionary power to influence humans sand, they reside within each of us, car- endemic in African apes. -

Movements of Ancient Human Endogenous Retroviruses
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.14.202135; this version posted July 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 2 Movements of ancient human endogenous retroviruses 3 detected in SOX2-expressing cells 4 5 6 Kazuaki Mondea,1, Yorifumi Satoub, Mizuki Gotoe, Yoshikazu Uchiyamac, Jumpei Itof, g, 7 Taku Kaitsukad, Hiromi Terasawaa, Shinya Yamagaa, Tomoya Matsusakoa, Fan-Yan 8 Weid, Ituro Inouef, Kazuhito Tomizawad, Akira Onoh, Takumi Erae, Tomohiro Sawaa, 9 Yosuke Maedaa 10 11 aDepartment of Microbiology, Faculty of Life Sciences, Kumamoto University, Chuo-ku, 12 860-8556, Kumamoto, Japan; bJoint Research Center for Human Retrovirus Infection, 13 Kumamoto University, Chuo-ku, 860-8556, Kumamoto, Japan; cDepartment of Medical 14 Physics, Kumamoto University, Chuo-ku, 862-0976, Kumamoto, Japan; dDepartment of 15 Molecular Physiology, Faculty of Life Sciences, Kumamoto University, Chuo-ku, 860-8556, 16 Kumamoto, Japan; eDepartment of Cell Modulation, Institute of Molecular Embryology and 17 Genetics (IMEG), Kumamoto University, Chuo-ku, 860-0811, Kumamoto, Japan; fNational 18 Institute of Genetics Laboratory of Human Genetics, Mishima, 411-8540, Shizuoka, Japan; 19 gDivision of Systems Virology, Department of infectious Disease Control, International 20 Research Center for infectious Diseases, Institute of Medical Science, The University of 21 Tokyo, 108-8639, Tokyo, Japan; hDepartment of Microbiology and Immunology, University 22 of Michigan Medical School, Ann Arbor, Michigan, 48109, United States of America.