Structural Basis for Spumavirus GAG Tethering to Chromatin
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Repression of Viral Gene Expression and Replication by the Unfolded Protein Response Effector Xbp1u Florian Hinte1, Eelco Van Anken2,3, Boaz Tirosh4, Wolfram Brune1*
RESEARCH ARTICLE Repression of viral gene expression and replication by the unfolded protein response effector XBP1u Florian Hinte1, Eelco van Anken2,3, Boaz Tirosh4, Wolfram Brune1* 1Heinrich Pette Institute, Leibniz Institute for Experimental Virology, Hamburg, Germany; 2Division of Genetics and Cell Biology, San Raffaele Scientific Institute, Milan, Italy; 3Universita` Vita-Salute San Raffaele, Milan, Italy; 4Institute for Drug Research, School of Pharmacy, Faculty of Medicine, The Hebrew University, Jerusalem, Israel Abstract The unfolded protein response (UPR) is a cellular homeostatic circuit regulating protein synthesis and processing in the ER by three ER-to-nucleus signaling pathways. One pathway is triggered by the inositol-requiring enzyme 1 (IRE1), which splices the X-box binding protein 1 (Xbp1) mRNA, thereby enabling expression of XBP1s. Another UPR pathway activates the activating transcription factor 6 (ATF6). Here we show that murine cytomegalovirus (MCMV), a prototypic b-herpesvirus, harnesses the UPR to regulate its own life cycle. MCMV activates the IRE1-XBP1 pathway early post infection to relieve repression by XBP1u, the product of the unspliced Xbp1 mRNA. XBP1u inhibits viral gene expression and replication by blocking the activation of the viral major immediate-early promoter by XBP1s and ATF6. These findings reveal a redundant function of XBP1s and ATF6 as activators of the viral life cycle, and an unexpected role of XBP1u as a potent repressor of both XBP1s and ATF6-mediated activation. *For correspondence: [email protected] Introduction The endoplasmic reticulum (ER) is responsible for synthesis, posttranslational modification, and fold- Competing interest: See ing of a substantial portion of cellular proteins. -

19-Kilodalton Tumor Antigen T
JOURNAL OF VIROLOGY, Nov. 1984, p. 336-343 Vol. 52, No. 2 0022-538X/84/110336-08$02.00/0 Copyright ©D 1984, American Society for Microbiology Adenovirus cyt+ Locus, Which Controls Cell Transformation and Tumorigenicity, Is an Allele of Ip+ Locus, Which Codes for a 19-Kilodalton Tumor Antigen T. SUBRAMANIAN,' MOHAN KUPPUSWAMY,1 STANLEY MAK,2 AND G. CHINNADURAI1* Institute for Molecular Virology, St. Louis University Medical Center, St. Louis, Missouri 63110,1 and Department of Biology, McMaster University, Hamilton, Ontario, Canada2 Received 30 April 1984/Accepted 19 July 1984 The early region Elb of adenovirus type 2 (Ad2) codes for two major tumor antigens of 53 and 19 kilodaltons (kd). The adenovirus Ip+ locus maps within the 19-kd tumor antigen-coding region (G. Chinnadurai, Cell 33:759-766, 1983). We have now constructed a large-plaque deletion mutant (d1250) of Ad2 that has a specific lesion in the 19-kd tumor antigen-coding region. In contrast to most other Ad2 lp mutants (G. Chinnadurai, Cell 33:759-766, 1983), mutant d1250 is cytocidal (cyt) on infected KB cells, causing extensive cellular destruction. Cells infected with Ad2 wt or most of these other Ad2 Ip mutants are rounded and aggregated without cell lysis (cyt+). The cyt phenotype of d1250 resembles the cyt mutants of highly oncogenic Adl2, isolated by Takemori et al. (Virology 36:575-586, 1968). By intertypic complementation analysis, we showed that the Adl2 cyt mutants indeed map within the 19-kd tumor antigen-coding region. The transforming potential of d1250 was assayed on an established rat embryo fibroblast cell line, CREF, and on primary rat embryo fibroblasts and baby rat kidney cells. -

Developing a Single-Cycle Infectious System to Study an ERV-K Retroviral Envelope
Developing a Single-Cycle Infectious System to Study an ERV-K Retroviral Envelope Author: Rana Elias Akleh Persistent link: http://hdl.handle.net/2345/bc-ir:107695 This work is posted on eScholarship@BC, Boston College University Libraries. Boston College Electronic Thesis or Dissertation, 2017 Copyright is held by the author. This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License (http:// creativecommons.org/licenses/by-nc-sa/4.0). Developing a Single-Cycle Infectious System to Study an ERV-K Retroviral Envelope Rana Elias Akleh A thesis submitted to the faculty of the department of Biology in partial fulfillment of the requirements for the degree of Master of Science Boston College Morrissey College of Arts and Sciences Graduate School September 2017 © 2017 Rana Elias Akleh Developing a Single-Cycle Infectious System to Study an ERV-K Retroviral Envelope Rana Elias Akleh Advisor: Welkin Johnson, Ph.D. Endogenous Retroviruses (ERVs) are “fossilized” retroviruses of a once exogenous retrovirus located in the genome of extant vertebrates. Retroviral infection results in a provirus integration into the host genome. An infection of a germline cell could lead to the provirus potentially being inherited by the offspring of the infected individual. Once in the genome, the provirus becomes subject to evolutionary processes and can become either lost or fixed in a population, remaining as “fossils” long after the exogenous retrovirus has gone extinct23. Notably, 8% of the human genome consists of ERVs30. Human Endogenous Retrovirus Type K (HERV-K)(HML-2) family is of particular interest. HERV-K integrations are as old as 30-35 million years, endogenizing before the separation of humans and Old World Monkeys. -

Ube1a Suppresses Herpes Simplex Virus-1 Replication
viruses Article UBE1a Suppresses Herpes Simplex Virus-1 Replication Marina Ikeda 1 , Akihiro Ito 2, Yuichi Sekine 1 and Masahiro Fujimuro 1,* 1 Department of Cell Biology, Kyoto Pharmaceutical University, Kyoto 607-8412, Japan; [email protected] (M.I.); [email protected] (Y.S.) 2 Laboratory of Cell Signaling, School of Life Sciences, Tokyo University of Pharmacy and Life Sciences, Tokyo 192-0392, Japan; [email protected] * Correspondence: [email protected]; Tel.: +81-75-595-4717 Academic Editors: Magdalena Weidner-Glunde and Andrea Lipi´nska Received: 7 November 2020; Accepted: 1 December 2020; Published: 4 December 2020 Abstract: Herpes simplex virus-1 (HSV-1) is the causative agent of cold sores, keratitis, meningitis, and encephalitis. HSV-1-encoded ICP5, the major capsid protein, is essential for capsid assembly during viral replication. Ubiquitination is a post-translational modification that plays a critical role in the regulation of cellular events such as proteasomal degradation, protein trafficking, and the antiviral response and viral events such as the establishment of infection and viral replication. Ub-activating enzyme (E1, also named UBE1) is involved in the first step in the ubiquitination. However, it is still unknown whether UBE1 contributes to viral infection or the cellular antiviral response. Here, we found that UBE1a suppressed HSV-1 replication and contributed to the antiviral response. The UBE1a inhibitor PYR-41 increased HSV-1 production. Immunofluorescence analysis revealed that UBE1a highly expressing cells presented low ICP5 expression, and vice versa. UBE1a inhibition by PYR-41 and shRNA increased ICP5 expression in HSV-1-infected cells. -

Researchers Unveil New Monkey Model for HIV 2 March 2009
Researchers unveil new monkey model for HIV 2 March 2009 By altering just one gene in HIV-1, scientists have evolving genes, APOBEC3 and TRIM5, which succeeded in infecting pig-tailed macaque produce unusual classes of defensive proteins with monkeys with a human version of the virus that distinctive capabilities to fight retroviruses such as has until now been impossible to study directly in HIV. These genes, shared by humans and their animals. The new strain of HIV has already been simian forebears, have evolved mutations specific used to demonstrate one method for preventing to each species' unique history of retroviral battles. infection and, with a little tweaking, could be a In most simians, the APOBEC3 and TRIM5 valuable model for vetting vaccine candidates. proteins actually kill HIV on sight, making it impossible for researchers to study the virus in an A team of researchers led by Paul Bieniasz and animal model. Instead, they have studied HIV's Theodora Hatziioannou at The Rockefeller cousin, simian immunodeficiency virus (SIV), which University showed that two pig-tailed macaques, causes an AIDS-like disease in certain monkey given a common antiretroviral treatment one week species. But SIV shares only about half of its amino before and one week after being exposed to the acid sequence with HIV, making it a very imperfect newly engineered HIV, had no signs of infection. substitute for testing anti-HIV drugs and vaccines. "We're not saying we can save the world with Several labs have engineered hybrids called SHIVs antiretroviral pills. But this model will allow us to — SIVs that contain pieces of HIV DNA — but these start studying the best way to administer have problematic differences, too. -

Chromatin Regulation of Virus Infection
Review TRENDS in Microbiology Vol.14 No.3 March 2006 Chromatin regulation of virus infection Paul M. Lieberman The Wistar Institute, Philadelphia, PA 19104, USA Cellular chromatin forms a dynamic structure that the position of the nucleosome on the DNA and its ability maintains the stability and accessibility of the host to form condensed higher-ordered structures such as DNA genome. Viruses that enter and persist in the heterochromatin can have profound effects on the acces- nucleus must, therefore, contend with the forces that sibility and activity of the packaged DNA. The ATP- drive chromatin formation and regulate chromatin dependent chromatin-remodeling complexes that contain structure. In some cases, cellular chromatin inhibits BRG1, BRM1 or SNF2h can alter the nucleosome viral gene expression and replication by suppressing positions and alter DNA accessibility to promote or DNA accessibility. In other cases, cellular chromatin repress transcription and replication [4]. Histone chaper- provides essential structure and organization to the viral ones and histone variants can also regulate chromosome genome and is necessary for successful completion of functions. For example, histone variant H2AX is phos- the viral life cycle. Consequently, viruses have acquired phorylated in response to DNA double-strand breaks and numerous mechanisms to manipulate cellular chroma- is thought to be essential for the recruitment of DNA tin to ensure viral genome survival and propagation. repair proteins [5]. The signaling mechanism and patterns of recognition, referred to as the histone code hypothesis, A chromatin perspective of virology can be markers for cellular division cycle and cancer Viruses are mobile genetic elements that must navigate prognosis [2,3,6]. -
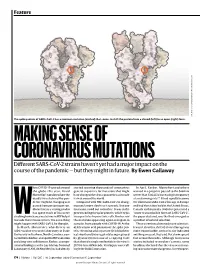
MAKING SENSE of CORONAVIRUS MUTATIONS Different SARS-Cov-2 Strains Haven’T Yet Had a Major Impact on the Course of the Pandemic — but They Might in Future
Feature SOURCE: STRUCTURAL DATA FROM K. SHEN & J. LUBAN K. SHEN FROM DATA STRUCTURAL SOURCE: The spike protein of SARS-CoV-2 has a common mutation (circled) that seems to shift the protein from a closed (left) to an open (right) form. MAKING SENSE OF CORONAVIRUS MUTATIONS Different SARS-CoV-2 strains haven’t yet had a major impact on the course of the pandemic — but they might in future. By Ewen Callaway hen COVID-19 spread around started scouring thousands of coronavirus In April, Korber, Montefiori and others the globe this year, David genetic sequences for mutations that might warned in a preprint posted to the bioRxiv Montefiori wondered how the have changed the virus’s properties as it made server that “D614G is increasing in frequency deadly virus behind the pan- its way around the world. at an alarming rate”1. It had rapidly become demic might be changing as it Compared with HIV, SARS-CoV-2 is chang- the dominant SARS-CoV-2 lineage in Europe passed from person to person. ing much more slowly as it spreads. But one and had then taken hold in the United States, Montefiori is a virologist who mutation stood out to Korber. It was in the Canada and Australia. D614G represented a has spent much of his career gene encoding the spike protein, which helps “more transmissible form of SARS-CoV-2”, Wstudying how chance mutations in HIV help it virus particles to penetrate cells. Korber saw the paper declared, one that had emerged as to evade the immune system. -
Croi 2021 Program Committee
General Information CONTENTS WELCOME . 2 General Information General Information OVERVIEW . 2 CONTINUING MEDICAL EDUCATION . 3 CONFERENCE SUPPORT . 4 VIRTUAL PLATFORM . 5 ON-DEMAND CONTENT AND WEBCASTS . 5 CONFERENCE SCHEDULE AT A GLANCE . 6 PRECONFERENCE SESSIONS . 9 LIVE PLENARY, ORAL, AND INTERACTIVE SESSIONS, AND ON-DEMAND SYMPOSIA BY DAY . 11 SCIENCE SPOTLIGHTS™ . 47 SCIENCE SPOTLIGHT™ SESSIONS BY CATEGORY . 109 CROI FOUNDATION . 112 IAS–USA . 112 CROI 2021 PROGRAM COMMITTEE . 113 Scientific Program Committee . 113 Community Liaison Subcommittee . 113 Former Members . 113 EXTERNAL REVIEWERS . .114 SCHOLARSHIP AWARDEES . 114 AFFILIATED OR PROXIMATE ACTIVITIES . 114 EMBARGO POLICIES AND SOCIAL MEDIA . 115 CONFERENCE ETIQUETTE . 115 ABSTRACT PROCESS Scientific Categories . 116 Abstract Content . 117 Presenter Responsibilities . 117 Abstract Review Process . 117 Statistics for Abstracts . 117 Abstracts Related to SARS-CoV-2 and Special Study Populations . 117. INDEX OF SPECIAL STUDY POPULATIONS . 118 INDEX OF PRESENTING AUTHORS . .122 . Version 9 .0 | Last Update on March 8, 2021 Printed in the United States of America . © Copyright 2021 CROI Foundation/IAS–USA . All rights reserved . ISBN #978-1-7320053-4-1 vCROI 2021 1 General Information WELCOME TO vCROI 2021 Welcome to vCROI 2021! The COVID-19 pandemic has changed the world for all of us in so many ways . Over the past year, we have had to put some of our HIV research on hold, learned to do our research in different ways using different tools, to communicate with each other in virtual formats, and to apply the many lessons in HIV research, care, and community advocacy to addressing the COVID-19 pandemic . Scientists and community stakeholders who have long been engaged in the endeavor to end the epidemic of HIV have pivoted to support and inform the unprecedented progress made in battle against SARS-CoV-2 . -

Final Scientific Programme
Final Scientific Programme Day 1 – Thursday, April 29 Arrival of conferees, registration, get-together party Session 1 Welcoming and Opening Remarks Chairman: Jan Svoboda 04:30 - 05:00 pm Jan Svoboda, Institute of Molecular Genetics, Prague Background and perspectives David Baltimore, California Institute of Technology, Pasadena 05:00 - 06:00 pm Keynote lecture 06:00 pm Welcome reception Day 2 – Friday, April 30 Session 2 Action at the Cell Surface Chairmen: John A.T. Young, Benjamin K. Chen 08:30 - 09:00 am John A.T. Young, Salk Institute, La Jolla Cellular factor involvement in retroviral replication 09:00 - 09:30 am Benjamin K. Chen, Mount Sinai School of Medicine, New York Spread of HIV through T cell virological synapses 09:30 - 09:45 am Paul Jolicoeur, Clinical Research Institute of Montreal Pathogenesis of HIV-1 Nef in thymocytes 09:45 - 10:00 am Michaela Rumlová, Institute of Organic Chemistry and Biochemistry, Prague Analysis of N-terminus of NC protein of M-PMV: its role in the particle assembly 10:00 - 10:30 am Coffee break Session 3 Identification of Essential Host Cell Factors Chairmen: Stephen P. Goff, Alan N. Engelmam 10:30 - 11:00 am Stephen P. Goff, Columbia University, New York HIV-1 Interactions with host proteins 11:00 - 11:30 am Alan N. Engelmam, Dana-Farber Cancer Institute, Boston HIV-1 preintegration complex function and host cell factors 11:30 - 11:45 am Peter Cherepanov, Imperial College London Structural basis for retroviral pre-integration complex assembly and strand transfer inhibitor action 11:45 - 12:00 am C. -

Heat Shock Protein 90 Chaperones E1A Early Protein of Adenovirus 5 and Is Essential for Replication of the Virus
International Journal of Molecular Sciences Article Heat Shock Protein 90 Chaperones E1A Early Protein of Adenovirus 5 and Is Essential for Replication of the Virus Iga Dalidowska 1, Olga Gazi 2, Dorota Sulejczak 1, Maciej Przybylski 2 and Pawel Bieganowski 1,* 1 Department of Experimental Pharmacology, Mossakowski Medical Research Institute, Polish Academy of Sciences, Pawinskiego 5, 02-106 Warsaw, Poland; [email protected] (I.D.); [email protected] (D.S.) 2 Chair and Department of Medical Microbiology, Medical University of Warsaw, 02-091 Warsaw, Poland; [email protected] (O.G.); [email protected] (M.P.) * Correspondence: [email protected] Abstract: Adenovirus infections tend to be mild, but they may pose a serious threat for young and immunocompromised individuals. The treatment is complicated because there are no approved safe and specific drugs for adenovirus infections. Here, we present evidence that 17-(Allylamino)-17- demethoxygeldanamycin (17-AAG), an inhibitor of Hsp90 chaperone, decreases the rate of human adenovirus 5 (HAdV-5) replication in cell cultures by 95%. 17-AAG inhibited the transcription of early and late genes of HAdV-5, replication of viral DNA, and expression of viral proteins. 6 h after infection, Hsp90 inhibition results in a 6.3-fold reduction of the newly synthesized E1A protein level without a decrease in the E1A mRNA level. However, the Hsp90 inhibition does not increase the decay rate of the E1A protein that was constitutively expressed in the cell before exposure to the inhibitor. The co-immunoprecipitation proved that E1A protein interacted with Hsp90. Altogether, the presented results show, for the first time. -

Rotavirus NSP1 Inhibits Nfkb Activation by Inducing Proteasome-Dependent Degradation of B-Trcp: a Novel Mechanism of IFN Antagonism
Rotavirus NSP1 Inhibits NFkB Activation by Inducing Proteasome-Dependent Degradation of b-TrCP: A Novel Mechanism of IFN Antagonism Joel W. Graff., Khalil Ettayebi., Michele E. Hardy* Veterinary Molecular Biology, Montana State University, Bozeman, Montana, United States of America Abstract Mechanisms by which viruses counter innate host defense responses generally involve inhibition of one or more components of the interferon (IFN) system. Multiple steps in the induction and amplification of IFN signaling are targeted for inhibition by viral proteins, and many of the IFN antagonists have direct or indirect effects on activation of latent cytoplasmic transcription factors. Rotavirus nonstructural protein NSP1 blocks transcription of type I IFNa/b by inducing proteasome-dependent degradation of IFN-regulatory factors 3 (IRF3), IRF5, and IRF7. In this study, we show that rotavirus NSP1 also inhibits activation of NFkB and does so by a novel mechanism. Proteasome-mediated degradation of inhibitor of kB(IkBa) is required for NFkB activation. Phosphorylated IkBa is a substrate for polyubiquitination by a multisubunit E3 ubiquitin ligase complex, Skp1/Cul1/F-box, in which the F-box substrate recognition protein is b-transducin repeat containing protein (b-TrCP). The data presented show that phosphorylated IkBa is stable in rotavirus-infected cells because infection induces proteasome-dependent degradation of b-TrCP. NSP1 expressed in isolation in transiently transfected cells is sufficient to induce this effect. Targeted degradation of an F-box protein of an E3 ligase complex with a prominent role in modulation of innate immune signaling and cell proliferation pathways is a unique mechanism of IFN antagonism and defines a second strategy of immune evasion used by rotaviruses. -

The Emerging Role of HIV-1 Integrase in Virion Morphogenesis
viruses Review Going beyond Integration: The Emerging Role of HIV-1 Integrase in Virion Morphogenesis Jennifer L. Elliott and Sebla B. Kutluay * Department of Molecular Microbiology, Washington University School of Medicine, Saint Louis, MO 63110, USA; [email protected] * Correspondence: [email protected] Received: 26 August 2020; Accepted: 7 September 2020; Published: 9 September 2020 Abstract: The HIV-1 integrase enzyme (IN) plays a critical role in the viral life cycle by integrating the reverse-transcribed viral DNA into the host chromosome. This function of IN has been well studied, and the knowledge gained has informed the design of small molecule inhibitors that now form key components of antiretroviral therapy regimens. Recent discoveries unveiled that IN has an under-studied yet equally vital second function in human immunodeficiency virus type 1 (HIV-1) replication. This involves IN binding to the viral RNA genome in virions, which is necessary for proper virion maturation and morphogenesis. Inhibition of IN binding to the viral RNA genome results in mislocalization of the viral genome inside the virus particle, and its premature exposure and degradation in target cells. The roles of IN in integration and virion morphogenesis share a number of common elements, including interaction with viral nucleic acids and assembly of higher-order IN multimers. Herein we describe these two functions of IN within the context of the HIV-1 life cycle, how IN binding to the viral genome is coordinated by the major structural protein, Gag, and discuss the value of targeting the second role of IN in virion morphogenesis. Keywords: HIV-1; integrase; maturation; integrase–RNA interactions; protein–RNA interactions 1.