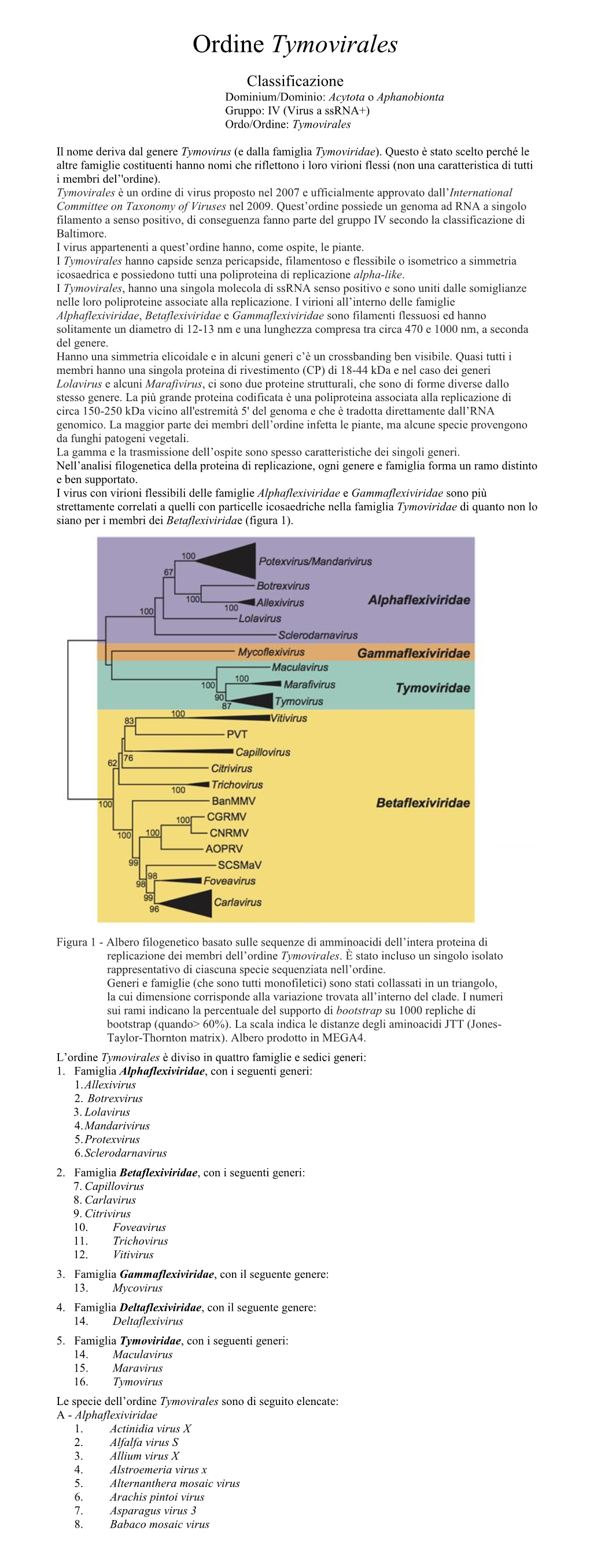
Ordine Tymovirales
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Examination of Potato Virus X Subgenomic Rnas: Their Umbn Er, Their Oc Ding Properties, and Their Possible Encapsidation
Louisiana State University LSU Digital Commons LSU Historical Dissertations and Theses Graduate School 1991 Examination of Potato Virus X Subgenomic RNAs: Their umbN er, Their oC ding Properties, and Their Possible Encapsidation. Mary A. Price Louisiana State University and Agricultural & Mechanical College Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_disstheses Recommended Citation Price, Mary A., "Examination of Potato Virus X Subgenomic RNAs: Their umbeN r, Their odC ing Properties, and Their osP sible Encapsidation." (1991). LSU Historical Dissertations and Theses. 5270. https://digitalcommons.lsu.edu/gradschool_disstheses/5270 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Historical Dissertations and Theses by an authorized administrator of LSU Digital Commons. For more information, please contact [email protected]. INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand corner and continuing from left to right in equal sections with small overlaps. -

Exploring the Tymovirids Landscape Through Metatranscriptomics Data
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.15.452586; this version posted July 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Exploring the tymovirids landscape through metatranscriptomics data 2 Nicolás Bejerman1,2, Humberto Debat1,2 3 4 1 Instituto de Patología Vegetal – Centro de Investigaciones Agropecuarias – Instituto Nacional de 5 Tecnología Agropecuaria (IPAVE-CIAP-INTA), Camino 60 Cuadras Km 5,5 (X5020ICA), Córdoba, 6 Argentina 7 2 Consejo Nacional de Investigaciones Científicas y Técnicas. Unidad de Fitopatología y Modelización 8 Agrícola, Camino 60 Cuadras Km 5,5 (X5020ICA), Córdoba, Argentina 9 10 Corresponding author: Nicolás Bejerman, [email protected] 11 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.07.15.452586; this version posted July 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 12 Abstract 13 Tymovirales is an order of viruses with positive-sense, single-stranded RNA genomes that mostly infect 14 plants, but also fungi and insects. The number of tymovirid sequences has been growing in the last few 15 years with the extensive use of high-throughput sequencing platforms. Here we report the discovery of 31 16 novel tymovirid genomes associated with 27 different host plant species, which were hidden in public 17 databases. -

Edna-Host: Detection of Global Plant Viromes Using High Throughput Sequencing
EDNA-HOST: DETECTION OF GLOBAL PLANT VIROMES USING HIGH THROUGHPUT SEQUENCING By LIZBETH DANIELA PENA-ZUNIGA Bachelor of Science in Biotechnology Escuela Politecnica de las Fuerzas Armadas (ESPE) Sangolqui, Ecuador 2014 Submitted to the Faculty of the Graduate College of the Oklahoma State University in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY May 2020 EDNA-HOST: DETECTION OF GLOBAL PLANT VIROMES USING HIGH THROUGHPUT SEQUENCING Dissertation Approved: Francisco Ochoa-Corona, Ph.D. Dissertation Adviser Committee member Akhtar, Ali, Ph.D. Committee member Hassan Melouk, Ph.D. Committee member Andres Espindola, Ph.D. Outside Committee Member Daren Hagen, Ph.D. ii ACKNOWLEDGEMENTS I would like to express sincere thanks to my major adviser Dr. Francisco Ochoa –Corona for his guidance from the beginning of my journey believing and trust that I am capable of developing a career as a scientist. I am thankful for his support and encouragement during hard times in research as well as in personal life. I truly appreciate the helpfulness of my advisory committee for their constructive input and guidance, thanks to: Dr. Akhtar Ali for his support in this research project and his kindness all the time, Dr. Hassan Melouk for his assistance, encouragement and his helpfulness in this study, Dr. Andres Espindola, developer of EDNA MiFi™, he was extremely helpful in every step of EDNA research, and for his willingness to give his time and advise; to Dr. Darren Hagen for his support and advise with bioinformatics and for his encouragement to develop a new set of research skills. I deeply appreciate Dr. -

Lily Virus X (LVX) ELISA Kit
Version 01-06/20 User's Manual Lily virus X (LVX) ELISA Kit DEIAPV181 2000T This product is for research use only and is not intended for diagnostic use. For illustrative purposes only. To perform the assay the instructions for use provided with the kit have to be used. Creative Diagnostics Address: 45-1 Ramsey Road, Shirley, NY 11967, USA Tel: 1-631-624-4882 (USA) 44-161-818-6441 (Europe) Fax: 1-631-938-8221 Email: [email protected] Web: www.creative-diagnostics.com Cat: DEIAPV181 Lily virus X (LVX) ELISA Kit Version 08-07/20 PRODUCT INFORMATION Intended Use The test can be used to detect LVX in host plants. General Description Lily virus X (LVX) is a pathogenic ssRNA(+) plant virus of the family Alphaflexiviridae and the order Tymovirales. It is the type species of the genus Potexvirus. LVX is able to infect all tissues of the lily plant. Virions have been cytopathologically detected in all parts of the host plants and contain approximately 5% nucleic acid and 95% protein, with no lipid contents. The mode of transmission is expected to be that of mechanical inoculation through insect vectors, since the spread of the virus was inhibited by insecticides but not mineral-oil sprays. Principles of Testing The enzyme-linked immunosorbent assay (ELISA) is a serological solid-phase method for identification of diseases based on antibodies and color change in the assay. In this method, specific antibodies are used to coat the microtitre plate, which then trap the target epitopes (antigens) from the viruses, bacteria and fungi. -

Blackberry Virus E: an Unusual flexivirus
Arch Virol (2011) 156:1665–1669 DOI 10.1007/s00705-011-1015-y BRIEF REPORT Blackberry virus E: an unusual flexivirus Sead Sabanadzovic • Nina Abou Ghanem-Sabanadzovic • Ioannis E. Tzanetakis Received: 5 November 2010 / Accepted: 30 April 2011 / Published online: 9 June 2011 Ó Springer-Verlag 2011 Abstract A virus, named blackberry virus E (BVE), was disease (BYVD), a serious disorder observed in the south- recently discovered in blackberries and characterized. The ern United States [19]. Disease symptoms are not specific to virus genome is 7,718 nt long, excluding the poly-A tail, any given virus combination, and different virus combina- contains five open reading frames (ORFs) and resembles that tions are found associated with identical symptoms [19]. of flexiviruses. Phylogenetic analysis revealed relationships This study was initiated with four blackberry plants to allexiviruses, which are known to infect plants of the showing BYVD-like symptoms observed in northeastern family Alliaceae. BVE lacks the 3’end-proximal ORF, Mississippi (Fig. 1A). They were tested by ELISA using which encodes a nucleotide-binding protein, a putative antibodies specific for 12 viruses and a ‘‘universal potyvirus’’ silencing suppressor in allexiviruses. The overall results of kit (Agdia Inc., USA). Additionally, they were tested by this study suggest that this virus is an atypical and as yet reverse transcription polymerase chain reaction (RT-PCR) undescribed flexivirus that is closely related to allexiviruses. using specific primers for viruses identified recently in BYVD-affected plants, or one still being characterized and Keywords Blackberry Á Virus Á dsRNA Á lacking serological diagnostics (authors, unpublished data). Alphaflexiviridae Á RT-PCR All four specimens were infected with blackberry virus Y, a virus known to be asymptomatic in single infections [17], leading to the assumption that one or more additional viruses Rubus species (blackberry, raspberry and their hybrids) are are involved in the observed symptomatology. -

Short 5′ UTR Enables Optimal Translation of Plant Virus Tricistronic RNA Via Leaky Scanning
bioRxiv preprint doi: https://doi.org/10.1101/2021.05.14.444105; this version posted May 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Title Short 5′ UTR enables optimal translation of plant virus tricistronic RNA via leaky scanning Authors Yuji Fujimoto1, Takuya Keima1, Masayoshi Hashimoto1, Yuka Hagiwara-Komoda2, Naoi Hosoe1, Shuko Nishida1, Takamichi Nijo1, Kenro Oshima3, Jeanmarie Verchot4, Shigetou Namba1, and Yasuyuki Yamaji1* 1Graduate School of Agricultural and Life Sciences, The University of Tokyo, 113-8657 Tokyo, Japan 2Department of Sustainable Agriculture, College of Agriculture, Food and Environment Sciences, Rakuno Gakuen University, 069-8501 Ebetsu Hokkaido, Japan 3Faculty of Bioscience, Hosei University, 3-7-2 Kajino-cho, Koganei-shi, 184-8584 Tokyo, Japan 4Department of Plant Pathology and Microbiology, Texas A&M University, College Station, TX 77802, USA. Correspondence *To whom correspondence should be addressed. Tel: +81-3-5841-5092. Fax: +81-3-5841-5090. E-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.05.14.444105; this version posted May 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Regardless of the general model of translation in eukaryotic cells, a number of studies suggested that many of mRNAs encode multiple proteins. -

Prediction of the Molecular Boundary and Evolutionary History of Novel Viral Alkb Domains Using Homology Modeling and Principal Component Analysis
Prediction of the Molecular Boundary and Evolutionary History of Novel Viral AlkB Domains Using Homology Modeling and Principal Component Analysis By Clayton Moore A Thesis presented to The University of Guelph In partial fulfilment of requirements for the degree of Master of Science in Bioinformatics Guelph, Ontario, Canada © Clayton Moore, September, 2018 ABSTRACT PREDICTION OF THE MOLECULAR BOUNDARY AND EVOLUTIONARY HISTORY OF NOVEL VIRAL ALKB DOMAINS USING HOMOLOGY MODELING AND PRINCIPAL COMPONENT ANALYSIS Clayton Moore Advisory Committee: University of Guelph, 2018 Dr. Baozhong Meng (Advisor) Dr. Jinzhong Fu Dr. Steffen Graether Dr. Lewis Lukens AlkB (Alkylation B) proteins are uBiquitous among cellular organisms where they act to reverse the damage in RNA and DNA due to methylation. The AlkB protein family is so significant to all forms of life that it was recently discovered that an AlkB domain is encoded as part of the replicase (poly)protein in a small suBset of single-stranded, positive-sense RNA viruses Belonging to five families. Interestingly, these AlkB-containing viruses are mostly important pathogens of perennials such as fruit crops. As a newly identified protein domain in RNA viruses, the origin, molecular Boundary of the viral AlkB domain as well as its function in viral replication and infection are unknown. The poor sequence conservation of this domain has made traditional Bioinformatic approaches unreliaBle and inconclusive. Here we apply principal component analysis, homology modelling, and traditional sequence Based techniques to propose a molecular Boundary and function to this novel viral domain. ACKNOWLEDGEMENTS This research project was supported By a grant from the Discovery Grant program (Project RGPIN-2014-05306) of the Natural Science and Engineering Research Council of Canada. -

Colour Break in Reverse Bicolour Daffodils Is Associated with the Presence of Narcissus Mosaic Virus Hunter Et Al
Colour break in reverse bicolour daffodils is associated with the presence of Narcissus mosaic virus Hunter et al. Hunter et al. Virology Journal 2011, 8:412 http://www.virologyj.com/content/8/1/412 (21 August 2011) Hunter et al. Virology Journal 2011, 8:412 http://www.virologyj.com/content/8/1/412 RESEARCH Open Access Colour break in reverse bicolour daffodils is associated with the presence of Narcissus mosaic virus Donald A Hunter1*, John D Fletcher2, Kevin M Davies1 and Huaibi Zhang1 Abstract Background: Daffodils (Narcissus pseudonarcissus) are one of the world’s most popular ornamentals. They also provide a scientific model for studying the carotenoid pigments responsible for their yellow and orange flower colours. In reverse bicolour daffodils, the yellow flower trumpet fades to white with age. The flowers of this type of daffodil are particularly prone to colour break whereby, upon opening, the yellow colour of the perianth is observed to be ‘broken’ into patches of white. This colour break symptom is characteristic of potyviral infections in other ornamentals such as tulips whose colour break is due to alterations in the presence of anthocyanins. However, reverse bicolour flowers displaying colour break show no other virus-like symptoms such as leaf mottling or plant stunting, leading some to argue that the carotenoid-based colour breaking in reverse bicolour flowers may not be caused by virus infection. Results: Although potyviruses have been reported to cause colour break in other flower species, enzyme-linked- immunoassays with an antibody specific to the potyviral family showed that potyviruses were not responsible for the occurrence of colour break in reverse bicolour daffodils. -

Molecular Characterization, Differential Movement and Construction of Infectious Cdna Clones of an Ohio Isolate of Hosta Virus X
Molecular characterization, differential movement and construction of infectious cDNA clones of an Ohio isolate of Hosta virus X Thesis Presented in Partial Fulfillment of the Requirements for The Degree Master of Science in the Graduate School of The Ohio State University By Carola De La Torre Cuba, BSc. The Ohio State University 2009 Thesis Committee: Dr. Dennis Lewandowski, Advisor Dr. Pierluigi Bonello Dr. Margaret G. Redinbaugh Copyright by Carola De La Torre 2009 Abstract Hostas (Hosta Tratt.) are the second most popular herbaceous perennial plant in the USA with sales exceeding $32 million in 2008 (USDA Floriculture crops 2008 Summary, 2009). Despite its relatively recent discovery in 1996, Hosta virus X (HVX) has already had a significant economic impact on hosta growers and producers. HVX is easily mechanically transmitted and can survive in the infected plants for years without showing symptoms. HVX has been reported in many states including Ohio. Very little is known about HVX biology with respect to diversity, symptom determinants and mechanisms of host resistance. The objectives of this research were to 1) Screen for resistance against HVX in a number of hosta cultivars, 2) Examine coat protein (CP) variability among Ohio isolates, 3) Molecular characterize an Ohio HVX isolate and 4) Construct an infectious cDNA clone of HVX. An Ohio HVX isolate, HVX-37, was selected for resistance screening, construction of an infectious cDNA clone and phylogenetic comparisons. Local and long-distance movement and accumulation of HVX-37 were determined for twenty-four hosta cultivars over three growing seasons resulting in five types of responses. Four cultivars (H. -

Hypera Postica) and Its Host Plant, Alfalfa (Medicago Sativa
Article Characterisation of the Viral Community Associated with the Alfalfa Weevil (Hypera postica) and Its Host Plant, Alfalfa (Medicago sativa) Sarah François 1,2,*, Aymeric Antoine-Lorquin 2, Maximilien Kulikowski 2, Marie Frayssinet 2, Denis Filloux 3,4, Emmanuel Fernandez 3,4, Philippe Roumagnac 3,4, Rémy Froissart 5 and Mylène Ogliastro 2,* 1 Peter Medawar Building for Pathogen Research, Department of Zoology, University of Oxford, South Park Road, Oxford OX1 3SY, UK 2 DGIMI Diversity, Genomes and Microorganisms–Insects Interactions, University of Montpellier, INRAE 34095 Montpellier, France; [email protected] (A.A.-L.); [email protected] (M.K.); [email protected] (M.F.) 3 CIRAD, UMR PHIM, 34090 Montpellier, France; [email protected] (D.F.); [email protected] (E.F.); [email protected] (P.R.) 4 PHIM Plant Health Institute, University of Montpellier, CIRAD, INRAE, Institut Agro, IRD 34090 Montpellier, France 5 MIVEGEC Infectious and Vector Diseases: Ecology, Genetics, Evolution and Control, University of Montpellier, CNRS, IRD 34394 Montpellier, France; [email protected] Citation: François, S.; * Correspondence: [email protected] (S.F.); [email protected] (M.O.) Antoine-Lorquin, A.; Kulikowski, M.; Frayssinet, M.; Abstract: Advances in viral metagenomics have paved the way of virus discovery by making the Filloux, D.; Fernandez, E.; exploration of viruses in any ecosystem possible. Applied to agroecosystems, such an approach Roumagnac, P.; Froissart, R.; opens new possibilities to explore how viruses circulate between insects and plants, which may help Ogliastro, M. Characterisation of the to optimise their management. It could also lead to identifying novel entomopathogenic viral re- Viral Community Associated with sources potentially suitable for biocontrol strategies. -

011P 2008.008P 2008.009P 2008.010P
Taxonomic proposal to the ICTV Executive Committee This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). 2008.008- Code(s) assigned: (to be completed by ICTV officers) 011P Short title: Creation of genus Lolavirus in the family Alphaflexiviridae for Lolium latent virus (e.g. 6 new species in the genus Zetavirus; re-classification of the family Zetaviridae etc.) Modules attached 1 2 3 4 5 (please check all that apply): 6 7 Author(s) with e-mail address(es) of the proposer: John Hammond ([email protected]) Anna Maria Vaira ([email protected]) Clarissa Maroon-Lango ([email protected]) ICTV-EC or Study Group comments and response of the proposer: The EC requested a phylogenetic tree that provided information on genetic distances between the sequences. This has been added to the proposal as Figure 3 by the chair of the plant virus subcommittee to further support the proposal. MODULE 4: NEW GENUS (if more than one genus is to be created, please complete additional copies of this section) Code 2008.008P (assigned by ICTV officers) To create a new genus assigned as follows: Subfamily: Fill in all that apply. Ideally, a genus should be placed within a higher taxon, Family: Alphaflexiviridae but if not put “unassigned” here. Order: Tymovirales Code 2008.009P (assigned by ICTV officers) To name the new genus: Lolavirus Code 2008.010P (assigned by ICTV officers) To assign the following as species in the new genus: Lolium latent virus -

The Characterisation of Ornithogalum Mosaic Virus
/ THE CHARACTERISATION OF ORNITHOGALUM MOSAIC VIRUS Johan Theodorus Burger University of Cape Town A dissertation submitted in fulfilment of the requiremements for the Degree of Doctor of Philosophy in the Faculty of Science, University of Cape Town. Cape Town, November, 1990 ·F-~~-=-~:.r:-":':."·.-:_~-~·;-.~--:-~·_::--:"_-.:-:::·_, ·_-:-... _... ';' _-:_: = -~-- -·-:--~ -~u 7 1! ~h: ~;;;-:f)~;_i~y :~(.\ ~~ ~~'·:~~ .. ~;.;s-~::;3:,_~:~:~~ '] c~v~1 ~:.:~.- -· . ·' -:~. ;: The copyright of this thesis vests in the author. No quotation from it or information derived from it is to be published without full acknowledgement of the source. The thesis is to be used for private study or non- commercial research purposes only. Published by the University of Cape Town (UCT) in terms of the non-exclusive license granted to UCT by the author. University of Cape Town CERTIFICATION OF SUPERVISOR' In terms of paragraph GP 8 in " General rules for the Degree of Doctor of Philosophy (PhD)", I as supervisor of the candidate, Johan Theodorus Burger, certify that I approve of- the incorporation in this dissertation of material that has already been submitted or accepted for publication. Associated Professor M. B. Von Wechmar Department of Microbiology University of Cape Town i CONTENTS ACKNOWLEDGEMENTS .................................... · ii ABBREVIATIONS . iii LIST OF FIGURES . vi LIST OFTABLES ................................ ; . ix ABSTRACT............................................ X CHAPTER 1: Introduction .......... "'· . 1 CHAPTER 2: The purification and physicochemical characterisation of ornithogalum mosaic virus. 33 CHAPTER 3: Serology of ornithogalum mosaic virus . 48 CHAPTER 4: Biological aspects of ornithogalum mosaic virus . 70 CHAPTER 5: Molecular cloning and nucleotide sequencing of ornithogalum mosaic virus. 88 CHAPTER 6: The expression of ornithogalum mosaic virus coat protein in E.