Novel Frameworks for Mining Heterogeneous and Dynamic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Interactive Proof Systems and Alternating Time-Space Complexity
Theoretical Computer Science 113 (1993) 55-73 55 Elsevier Interactive proof systems and alternating time-space complexity Lance Fortnow” and Carsten Lund** Department of Computer Science, Unicersity of Chicago. 1100 E. 58th Street, Chicago, IL 40637, USA Abstract Fortnow, L. and C. Lund, Interactive proof systems and alternating time-space complexity, Theoretical Computer Science 113 (1993) 55-73. We show a rough equivalence between alternating time-space complexity and a public-coin interactive proof system with the verifier having a polynomial-related time-space complexity. Special cases include the following: . All of NC has interactive proofs, with a log-space polynomial-time public-coin verifier vastly improving the best previous lower bound of LOGCFL for this model (Fortnow and Sipser, 1988). All languages in P have interactive proofs with a polynomial-time public-coin verifier using o(log’ n) space. l All exponential-time languages have interactive proof systems with public-coin polynomial-space exponential-time verifiers. To achieve better bounds, we show how to reduce a k-tape alternating Turing machine to a l-tape alternating Turing machine with only a constant factor increase in time and space. 1. Introduction In 1981, Chandra et al. [4] introduced alternating Turing machines, an extension of nondeterministic computation where the Turing machine can make both existential and universal moves. In 1985, Goldwasser et al. [lo] and Babai [l] introduced interactive proof systems, an extension of nondeterministic computation consisting of two players, an infinitely powerful prover and a probabilistic polynomial-time verifier. The prover will try to convince the verifier of the validity of some statement. -

ACM Honors Eminent Researchers for Technical Innovations That Have Improved How We Live and Work 2016 Recipients Made Contribu
Contact: Jim Ormond 212-626-0505 [email protected] ACM Honors Eminent Researchers for Technical Innovations That Have Improved How We Live and Work 2016 Recipients Made Contributions in Areas Including Big Data Analysis, Computer Vision, and Encryption NEW YORK, NY, May 3, 2017 – ACM, the Association for Computing Machinery (www.acm.org), today announced the recipients of four prestigious technical awards. These leaders were selected by their peers for making significant contributions that have had far-reaching impact on how we live and work. The awards reflect achievements in file sharing, broadcast encryption, information visualization and computer vision. The 2016 recipients will be formally honored at the ACM Awards Banquet on June 24 in San Francisco. The 2016 Award Recipients include: • Mahadev Satyanarayanan, Michael L. Kazar, Robert N. Sidebotham, David A. Nichols, Michael J. West, John H. Howard, Alfred Z. Spector and Sherri M. Nichols, recipients of the ACM Software System Award for developing the Andrew File System (AFS). AFS was the first distributed file system designed for tens of thousands of machines, and pioneered the use of scalable, secure and ubiquitous access to shared file data. To achieve the goal of providing a common shared file system used by large networks of people, AFS introduced novel approaches to caching, security, management and administration. AFS is still in use today as both an open source system and as the file system in commercial applications. It has also inspired several cloud-based storage applications. The 2016 Software System Award recipients designed and built the Andrew File System in the 1980s while working as a team at the Information Technology Center (ITC), a partnership between Carnegie Mellon University and IBM. -

Compact E-Cash and Simulatable Vrfs Revisited
Compact E-Cash and Simulatable VRFs Revisited Mira Belenkiy1, Melissa Chase2, Markulf Kohlweiss3, and Anna Lysyanskaya4 1 Microsoft, [email protected] 2 Microsoft Research, [email protected] 3 KU Leuven, ESAT-COSIC / IBBT, [email protected] 4 Brown University, [email protected] Abstract. Efficient non-interactive zero-knowledge proofs are a powerful tool for solving many cryptographic problems. We apply the recent Groth-Sahai (GS) proof system for pairing product equations (Eurocrypt 2008) to two related cryptographic problems: compact e-cash (Eurocrypt 2005) and simulatable verifiable random functions (CRYPTO 2007). We present the first efficient compact e-cash scheme that does not rely on a ran- dom oracle. To this end we construct efficient GS proofs for signature possession, pseudo randomness and set membership. The GS proofs for pseudorandom functions give rise to a much cleaner and substantially faster construction of simulatable verifiable random functions (sVRF) under a weaker number theoretic assumption. We obtain the first efficient fully simulatable sVRF with a polynomial sized output domain (in the security parameter). 1 Introduction Since their invention [BFM88] non-interactive zero-knowledge proofs played an important role in ob- taining feasibility results for many interesting cryptographic primitives [BG90,GO92,Sah99], such as the first chosen ciphertext secure public key encryption scheme [BFM88,RS92,DDN91]. The inefficiency of these constructions often motivated independent practical instantiations that were arguably conceptually less elegant, but much more efficient ([CS98] for chosen ciphertext security). We revisit two important cryptographic results of pairing-based cryptography, compact e-cash [CHL05] and simulatable verifiable random functions [CL07], that have very elegant constructions based on non-interactive zero-knowledge proof systems, but less elegant practical instantiations. -

Information Theory Methods in Communication Complexity
INFORMATION THEORY METHODS IN COMMUNICATION COMPLEXITY BY NIKOLAOS LEONARDOS A dissertation submitted to the Graduate School—New Brunswick Rutgers, The State University of New Jersey in partial fulfillment of the requirements for the degree of Doctor of Philosophy Graduate Program in Computer Science Written under the direction of Michael Saks and approved by New Brunswick, New Jersey JANUARY, 2012 ABSTRACT OF THE DISSERTATION Information theory methods in communication complexity by Nikolaos Leonardos Dissertation Director: Michael Saks This dissertation is concerned with the application of notions and methods from the field of information theory to the field of communication complexity. It con- sists of two main parts. In the first part of the dissertation, we prove lower bounds on the random- ized two-party communication complexity of functions that arise from read-once boolean formulae. A read-once boolean formula is a formula in propositional logic with the property that every variable appears exactly once. Such a formula can be represented by a tree, where the leaves correspond to variables, and the in- ternal nodes are labeled by binary connectives. Under certain assumptions, this representation is unique. Thus, one can define the depth of a formula as the depth of the tree that represents it. The complexity of the evaluation of general read-once formulae has attracted interest mainly in the decision tree model. In the communication complexity model many interesting results deal with specific read-once formulae, such as disjointness and tribes. In this dissertation we use information theory methods to prove lower bounds that hold for any read-once ii formula. -

ANDREAS PIERIS School of Informatics, University of Edinburgh 10 Crichton Street, Edinburgh, EH8 9AB, UK [email protected]
ANDREAS PIERIS School of Informatics, University of Edinburgh 10 Crichton Street, Edinburgh, EH8 9AB, UK [email protected] UNIVERSITY EDUCATION • D.Phil. in Computer Science, 2011 Department of Computer Science, University of Oxford Thesis: Ontological Query Answering: New Languages, Algorithms and Complexity Supervisor: Professor Georg Gottlob • M.Sc. in Mathematics anD FounDations oF Computer Science (with Distinction), 2007 Mathematical Institute, University of Oxford Thesis: Data Exchange and Schema Mappings Supervisor: Professor Georg Gottlob • B.Sc. in Computer Science (with Distinction, GPA: 9.06/10), 2006 Department of Computer Science, University of Cyprus Thesis: The Fully Mixed Nash Equilibrium Conjecture Supervisor: Professor Marios Mavronicolas EMPLOYMENT HISTORY • Lecturer (equivalent to Assistant ProFessor) in Databases, 09/2016 – present School of Informatics, University of Edinburgh • PostDoctoral Researcher, 11/2014 – 09/2016 Institute of Logic and Computation, Vienna University of Technology • PostDoctoral Researcher, 09/2011 – 10/2014 Department of Computer Science, University of Oxford RESEARCH Major research interests • Data management: knowledge-enriched data, uncertain data • Knowledge representation and reasoning: ontology languages, complexity of reasoning • Computational logic and its applications to computer science Research grants • EfFicient Querying oF Inconsistent Data, 09/2018 – 08/2022 Principal Investigator Funding agency: Engineering and Physical Sciences Research Council (EPSRC) Total award: £758,049 • Value AdDeD Data Systems: Principles anD Architecture, 04/2015 – 03/2020 Co-Investigator Funding agency: Engineering and Physical Sciences Research Council (EPSRC) Total award: £1,546,471 Research supervision experience • Marco Calautti, postdoctoral supervision, University of Edinburgh, 09/2016 – present • Markus Schneider, Ph.D. supervisor, University of Edinburgh, 09/2018 – present • Gerald Berger, Ph.D. -

UNIVERSITY of CALIFORNIA RIVERSIDE Unsupervised And
UNIVERSITY OF CALIFORNIA RIVERSIDE Unsupervised and Zero-Shot Learning for Open-Domain Natural Language Processing A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Computer Science by Muhammad Abu Bakar Siddique June 2021 Dissertation Committee: Dr. Evangelos Christidis, Chairperson Dr. Amr Magdy Ahmed Dr. Samet Oymak Dr. Evangelos Papalexakis Copyright by Muhammad Abu Bakar Siddique 2021 The Dissertation of Muhammad Abu Bakar Siddique is approved: Committee Chairperson University of California, Riverside To my family for their unconditional love and support. i ABSTRACT OF THE DISSERTATION Unsupervised and Zero-Shot Learning for Open-Domain Natural Language Processing by Muhammad Abu Bakar Siddique Doctor of Philosophy, Graduate Program in Computer Science University of California, Riverside, June 2021 Dr. Evangelos Christidis, Chairperson Natural Language Processing (NLP) has yielded results that were unimaginable only a few years ago on a wide range of real-world tasks, thanks to deep neural networks and the availability of large-scale labeled training datasets. However, existing supervised methods assume an unscalable requirement that labeled training data is available for all classes: the acquisition of such data is prohibitively laborious and expensive. Therefore, zero-shot (or unsupervised) models that can seamlessly adapt to new unseen classes are indispensable for NLP methods to work in real-world applications effectively; such models mitigate (or eliminate) the need for collecting and annotating data for each domain. This dissertation ad- dresses three critical NLP problems in contexts where training data is scarce (or unavailable): intent detection, slot filling, and paraphrasing. Having reliable solutions for the mentioned problems in the open-domain setting pushes the frontiers of NLP a step towards practical conversational AI systems. -

Handbook of Formal Languages
Handbook of Formal Languages von Grzegorz Rozenberg, Arto Salomaa 1. Auflage Handbook of Formal Languages – Rozenberg / Salomaa schnell und portofrei erhältlich bei beck-shop.de DIE FACHBUCHHANDLUNG Springer 1997 Verlag C.H. Beck im Internet: www.beck.de ISBN 978 3 540 60420 4 Contents of Volume 1 Chapter 1. Formal Languages: an Introduction and a Synopsis Alexandru Mateescu and Arto Salomaa ............................ 1 1. Languages, formal and natural ................................. 1 1.1 Historical linguistics ...................................... 3 1.2 Language and evolution ................................... 7 1.3 Language and neural structures ............................ 9 2. Glimpses of mathematical language theory ...................... 9 2.1 Words and languages ..................................... 10 2.2 About commuting ........................................ 12 2.3 About stars ............................................. 15 2.4 Avoiding scattered subwords .............................. 19 2.5 About scattered residuals ................................. 24 3. Formal languages: a telegraphic survey ......................... 27 3.1 Language and grammar. Chomsky hierarchy ................. 27 3.2 Regular and context-free languages ......................... 31 3.3 L Systems............................................... 33 3.4 More powerful grammars and grammar systems .............. 35 3.5 Books on formal languages ................................ 36 References ..................................................... 38 Chapter -
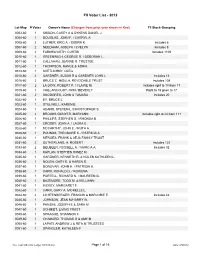
FII Voter Ledger
FII Voter List - 2012 Lot-Map # Votes Owner's Name (Changes from prior year shown in Red) FII Stock Grouping 0003-60 1 SISSON, CAREY A & DYKENS DANIEL J 0004-60 1 DOUGLAS, JOHN F. / CHERYL A. 0005-60 2 LUTHER, ERIC A. / DEBRA K. Includes 6 0007-60 2 NEEDHAM, JOSEPH / EVELYN Includes 8 0009-60 2 FARNSWORTH, CURTIS Includes 1109 0010-60 1 GREENWICH, GEORGE R. / DEBORAH L. 0011-60 1 CALLAHAN, JOANNE R. TRUSTEE 0012-60 1 THOMPSON, RANCE & ADRIA 0013-60 1 SVETLICHNY, OLEG 0015-60 2 GARDNER, SUSAN B & GARDNER JOHN L Includes 14 0016-60 2 BRUCE C. NISULA, REVOCABLE TRUST Includes 108 0017-60 2 LA DOW, ROBERT P. / CLAIRE M. Includes right to 18 from 17 0019-60 1 VAILLANCOURT, RON / BEVERLY Right to 18 given to 17 0021-60 2 SNODGRES, JOHN & TAMARA Includes 20 0022-60 1 EY, BRUCE L. 0023-60 1 STILLWELL, KAREN B. 0024-60 1 ADAMS, STEVEN L./CHRISTOPHER S. 0025-60 2 BROOKS-GONYER, MARYANN Includes right to 24 from 111 0026-60 1 PHILLIPS, STEPHEN S. / RHONDA B. 0027-60 1 CROSBY, JOHN A. / LAURA E. 0028-60 1 MCCARTHY, JOHN E. / RUTH A. 0029-60 1 POUNDS, THEODORE K. / PATRICIA A. 0030-60 1 MERGES, FRANK & AEJA FAMILY TRUST 0031-60 2 SUTHERLAND, H. ROBERT Includes 122 0033-60 2 DEUBLER, RUSSELL A. / MARCIA A. Includes 32 0034-60 1 KAPLAN, STEPHEN H/INEZ M 0035-60 1 GARDNER, KENNETH R. & NOLEN KATHLEEN L. 0036-60 1 NOLEN, GARY E. & KAREN R. 0037-60 1 DONOVAN, JOHN H. -

On SZK and PP
Electronic Colloquium on Computational Complexity, Revision 2 of Report No. 140 (2016) On SZK and PP Adam Bouland1, Lijie Chen2, Dhiraj Holden1, Justin Thaler3, and Prashant Nalini Vasudevan1 1CSAIL, Massachusetts Institute of Technology, Cambridge, MA USA 2IIIS, Tsinghua University, Beijing, China 3Georgetown University, Washington, DC USA Abstract In both query and communication complexity, we give separations between the class NISZK, con- taining those problems with non-interactive statistical zero knowledge proof systems, and the class UPP, containing those problems with randomized algorithms with unbounded error. These results significantly improve on earlier query separations of Vereschagin [Ver95] and Aaronson [Aar12] and earlier commu- nication complexity separations of Klauck [Kla11] and Razborov and Sherstov [RS10]. In addition, our results imply an oracle relative to which the class NISZK 6⊆ PP. This answers an open question of Wa- trous from 2002 [Aar]. The technical core of our result is a stronger hardness amplification theorem for approximate degree, which roughly says that composing the gapped-majority function with any function of high approximate degree yields a function with high threshold degree. Using our techniques, we also give oracles relative to which the following two separations hold: perfect zero knowledge (PZK) is not contained in its complement (coPZK), and SZK (indeed, even NISZK) is not contained in PZK (indeed, even HVPZK). Along the way, we show that HVPZK is contained in PP in a relativizing manner. We prove a number of implications of these results, which may be of independent interest outside of structural complexity. Specifically, our oracle separation implies that certain parameters of the Polariza- tion Lemma of Sahai and Vadhan [SV03] cannot be much improved in a black-box manner. -

FOCS 2005 Program SUNDAY October 23, 2005
FOCS 2005 Program SUNDAY October 23, 2005 Talks in Grand Ballroom, 17th floor Session 1: 8:50am – 10:10am Chair: Eva´ Tardos 8:50 Agnostically Learning Halfspaces Adam Kalai, Adam Klivans, Yishay Mansour and Rocco Servedio 9:10 Noise stability of functions with low influences: invari- ance and optimality The 46th Annual IEEE Symposium on Elchanan Mossel, Ryan O’Donnell and Krzysztof Foundations of Computer Science Oleszkiewicz October 22-25, 2005 Omni William Penn Hotel, 9:30 Every decision tree has an influential variable Pittsburgh, PA Ryan O’Donnell, Michael Saks, Oded Schramm and Rocco Servedio Sponsored by the IEEE Computer Society Technical Committee on Mathematical Foundations of Computing 9:50 Lower Bounds for the Noisy Broadcast Problem In cooperation with ACM SIGACT Navin Goyal, Guy Kindler and Michael Saks Break 10:10am – 10:30am FOCS ’05 gratefully acknowledges financial support from Microsoft Research, Yahoo! Research, and the CMU Aladdin center Session 2: 10:30am – 12:10pm Chair: Satish Rao SATURDAY October 22, 2005 10:30 The Unique Games Conjecture, Integrality Gap for Cut Problems and Embeddability of Negative Type Metrics Tutorials held at CMU University Center into `1 [Best paper award] Reception at Omni William Penn Hotel, Monongahela Room, Subhash Khot and Nisheeth Vishnoi 17th floor 10:50 The Closest Substring problem with small distances Tutorial 1: 1:30pm – 3:30pm Daniel Marx (McConomy Auditorium) Chair: Irit Dinur 11:10 Fitting tree metrics: Hierarchical clustering and Phy- logeny Subhash Khot Nir Ailon and Moses Charikar On the Unique Games Conjecture 11:30 Metric Embeddings with Relaxed Guarantees Break 3:30pm – 4:00pm Ittai Abraham, Yair Bartal, T-H. -

Interactive Proofs
Interactive proofs April 12, 2014 [72] L´aszl´oBabai. Trading group theory for randomness. In Proc. 17th STOC, pages 421{429. ACM Press, 1985. doi:10.1145/22145.22192. [89] L´aszl´oBabai and Shlomo Moran. Arthur-Merlin games: A randomized proof system and a hierarchy of complexity classes. J. Comput. System Sci., 36(2):254{276, 1988. doi:10.1016/0022-0000(88)90028-1. [99] L´aszl´oBabai, Lance Fortnow, and Carsten Lund. Nondeterministic ex- ponential time has two-prover interactive protocols. In Proc. 31st FOCS, pages 16{25. IEEE Comp. Soc. Press, 1990. doi:10.1109/FSCS.1990.89520. See item 1991.108. [108] L´aszl´oBabai, Lance Fortnow, and Carsten Lund. Nondeterministic expo- nential time has two-prover interactive protocols. Comput. Complexity, 1 (1):3{40, 1991. doi:10.1007/BF01200056. Full version of 1990.99. [136] Sanjeev Arora, L´aszl´oBabai, Jacques Stern, and Z. (Elizabeth) Sweedyk. The hardness of approximate optima in lattices, codes, and systems of linear equations. In Proc. 34th FOCS, pages 724{733, Palo Alto CA, 1993. IEEE Comp. Soc. Press. doi:10.1109/SFCS.1993.366815. Conference version of item 1997:160. [160] Sanjeev Arora, L´aszl´oBabai, Jacques Stern, and Z. (Elizabeth) Sweedyk. The hardness of approximate optima in lattices, codes, and systems of linear equations. J. Comput. System Sci., 54(2):317{331, 1997. doi:10.1006/jcss.1997.1472. Full version of 1993.136. [111] L´aszl´oBabai, Lance Fortnow, Noam Nisan, and Avi Wigderson. BPP has subexponential time simulations unless EXPTIME has publishable proofs. In Proc. -

UC Irvine UC Irvine Previously Published Works
UC Irvine UC Irvine Previously Published Works Title The capacity of feedforward neural networks. Permalink https://escholarship.org/uc/item/29h5t0hf Authors Baldi, Pierre Vershynin, Roman Publication Date 2019-08-01 DOI 10.1016/j.neunet.2019.04.009 License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California THE CAPACITY OF FEEDFORWARD NEURAL NETWORKS PIERRE BALDI AND ROMAN VERSHYNIN Abstract. A long standing open problem in the theory of neural networks is the devel- opment of quantitative methods to estimate and compare the capabilities of different ar- chitectures. Here we define the capacity of an architecture by the binary logarithm of the number of functions it can compute, as the synaptic weights are varied. The capacity provides an upperbound on the number of bits that can be extracted from the training data and stored in the architecture during learning. We study the capacity of layered, fully-connected, architectures of linear threshold neurons with L layers of size n1, n2,...,nL and show that in essence the capacity is given by a cubic polynomial in the layer sizes: L−1 C(n1,...,nL) = Pk=1 min(n1,...,nk)nknk+1, where layers that are smaller than all pre- vious layers act as bottlenecks. In proving the main result, we also develop new techniques (multiplexing, enrichment, and stacking) as well as new bounds on the capacity of finite sets. We use the main result to identify architectures with maximal or minimal capacity under a number of natural constraints.