A Genetic System for Methanocaldococcus Jannaschii
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -

Genomics and Energy and Environmental Science Poster 2011
Omics: Exploring the Molecular Universe within Biological Systems Instead of studying one or a few genes or proteins at a time, “omics” collectively Proteomics is the analysis of the proteome—the complete set describes the comprehensive analysis of genes, RNA transcripts, proteins, of proteins expressed by a cell or population of cells. Proteins, metabolites, and other molecules present in a biological system. Ongoing the workhorse molecules of life, catalyze biochemical reactions; advances in computing power and automated technologies for DNA sequencing provide structural support; and recognize, bind, or transport and experiments continue to improve our ability to analyze increasing numbers other molecules throughout the cell. Hundreds of dierent types of molecules and how they function as a system. Systems biology integrates the of proteins can be expressed at a time, and most are part of large data from various omic analyses using computational tools to build predictive complexes made up of models of biological systems. many proteins and other molecules. Transcriptomics is the analysis of the transcriptome—the complete set of RNA Hundreds to molecules present in a cell or population of thousands of different cells. RNA, which is much less stable than protein types exist within DNA, is constantly being synthesized and a cell. This large subunit of a ribosome contains about then broken down to facilitate rapid 3,000 RNA nucleotides changes in paerns of protein expression (gray) and 30 protein that occur as an organism dynamically chains (gold). responds to its environment. In addition to the three major classes of RNA (mRNA, tRNA, and ribosomal RNA), single- stranded RNA is very exible and can fold into complex shapes that carry out specic functions. -

Biological Diversity in the Patent System
Biological Diversity in the Patent System Paul Oldham1,2*, Stephen Hall1,3, Oscar Forero1,4 1 ESRC Centre for Economic and Social Aspects of Genomics (Cesagen), Lancaster University, Lancaster, United Kingdom, 2 Institute of Advanced Studies, United Nations University, Yokohama, Japan, 3 One World Analytics, Lancaster, United Kingdom, 4 Centre for Development, Environment and Policy, SOAS, University of London, London, United Kingdom Abstract Biological diversity in the patent system is an enduring focus of controversy but empirical analysis of the presence of biodiversity in the patent system has been limited. To address this problem we text mined 11 million patent documents for 6 million Latin species names from the Global Names Index (GNI) established by the Global Biodiversity Information Facility (GBIF) and Encyclopedia of Life (EOL). We identified 76,274 full Latin species names from 23,882 genera in 767,955 patent documents. 25,595 species appeared in the claims section of 136,880 patent documents. This reveals that human innovative activity involving biodiversity in the patent system focuses on approximately 4% of taxonomically described species and between 0.8–1% of predicted global species. In this article we identify the major features of the patent landscape for biological diversity by focusing on key areas including pharmaceuticals, neglected diseases, traditional medicines, genetic engineering, foods, biocides, marine genetic resources and Antarctica. We conclude that the narrow focus of human innovative activity and ownership of genetic resources is unlikely to be in the long term interest of humanity. We argue that a broader spectrum of biodiversity needs to be opened up to research and development based on the principles of equitable benefit-sharing, respect for the objectives of the Convention on Biological Diversity, human rights and ethics. -

A Virus That Infects a Hyperthermophile Encapsidates A-Form
RESEARCH | REPORTS we observe sets of regulatory sites that exhibit Illumina, Inc. One or more embodiments of one or more patents SUPPLEMENTARY MATERIALS patterns of coordinated regulation (e.g., LYN, and patent applications filed by Illumina may encompass the www.sciencemag.org/content/348/6237/910/suppl/DC1 encoding a tyrosine kinase involved in B cell methods, reagents, and data disclosed in this manuscript. All Materials and Methods methods for making the transposase complexes are described in signaling) (Fig. 4B), although reproducibility of Figs. S1 to S22 (18); however, Illumina will provide transposase complexes in Tables S1 and S2 these patterns across biological replicates was response to reasonable requests from the scientific community References (24–39) modest (fig. S22). Given the sparsity of the data, subject to a material transfer agreement. Some work in this study identifying pairs of coaccessible DNA elements is related to technology described in patent applications 19 March 2015; accepted 24 April 2015 WO2014142850, 2014/0194324, 2010/0120098, 2011/0287435, Published online 7 May 2015; within individual loci is statistically challenging 2013/0196860, and 2012/0208705. 10.1126/science.aab1601 and merits further development. We report chromatin accessibility maps for >15,000 single cells. Our combinatorial cellular indexing scheme could feasibly be scaled to col- VIROLOGY lect data from ~17,280 cells per experiment by using 384-by-384 barcoding and sorting 100 nu- clei per well (assuming similar cell recovery and A virus that infects a collision rates) (fig. S1) (19). Particularly as large- scale efforts to build a human cell atlas are con- templated (23), it is worth noting that because hyperthermophile encapsidates DNA is at uniform copy number, single-cell chro- matin accessibility mapping may require far fewer A-form DNA reads per single cell to define cell types, relative to single-cell RNA-seq. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Archaeology of Eukaryotic DNA Replication
Downloaded from http://cshperspectives.cshlp.org/ on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Archaeology of Eukaryotic DNA Replication Kira S. Makarova and Eugene V. Koonin National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland 20894 Correspondence: [email protected] Recent advances in the characterization of the archaeal DNA replication system together with comparative genomic analysis have led to the identification of several previously un- characterized archaeal proteins involved in replication and currently reveal a nearly com- plete correspondence between the components of the archaeal and eukaryotic replication machineries. It can be inferred that the archaeal ancestor of eukaryotes and even the last common ancestor of all extant archaea possessed replication machineries that were compa- rable in complexity to the eukaryotic replication system. The eukaryotic replication system encompasses multiple paralogs of ancestral components such that heteromeric complexes in eukaryotes replace archaeal homomeric complexes, apparently along with subfunctionali- zation of the eukaryotic complex subunits. In the archaea, parallel, lineage-specific dupli- cations of many genes encoding replication machinery components are detectable as well; most of these archaeal paralogs remain to be functionally characterized. The archaeal rep- lication system shows remarkable plasticity whereby even some essential components such as DNA polymerase and single-stranded DNA-binding protein are displaced by unrelated proteins with analogous activities in some lineages. ouble-stranded DNA is the molecule that Okazaki fragments (Kornberg and Baker 2005; Dcarries genetic information in all cellular Barry and Bell 2006; Hamdan and Richardson life-forms; thus, replication of this genetic ma- 2009; Hamdan and van Oijen 2010). -

Extremely Thermophilic Microorganisms As Metabolic Engineering Platforms for Production of Fuels and Industrial Chemicals
REVIEW published: 05 November 2015 doi: 10.3389/fmicb.2015.01209 Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals Benjamin M. Zeldes 1, Matthew W. Keller 2, Andrew J. Loder 1, Christopher T. Straub 1, Michael W. W. Adams 2 and Robert M. Kelly 1* 1 Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, NC, USA, 2 Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA, USA Enzymes from extremely thermophilic microorganisms have been of technological interest for some time because of their ability to catalyze reactions of industrial significance at elevated temperatures. Thermophilic enzymes are now routinely produced in recombinant mesophilic hosts for use as discrete biocatalysts. Genome and metagenome sequence data for extreme thermophiles provide useful information for putative biocatalysts for a wide range of biotransformations, albeit involving at most a few enzymatic steps. However, in the past several years, unprecedented progress has been made in establishing molecular genetics tools for extreme thermophiles to the point Edited by: that the use of these microorganisms as metabolic engineering platforms has become Bettina Siebers, University of Duisburg-Essen, possible. While in its early days, complex metabolic pathways have been altered or Germany engineered into recombinant extreme thermophiles, such that the production of fuels and Reviewed by: chemicals at elevated temperatures has become possible. Not only does this expand the Haruyuki Atomi, thermal range for industrial biotechnology, it also potentially provides biodiverse options Kyoto University, Japan Phillip Craig Wright, for specific biotransformations unique to these microorganisms. The list of extreme University of Sheffield, UK thermophiles growing optimally between 70 and 100◦C with genetic toolkits currently *Correspondence: available includes archaea and bacteria, aerobes and anaerobes, coming from genera Robert M. -
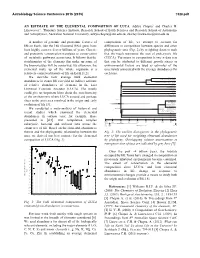
An Estimate of the Elemental Composition of Luca
Astrobiology Science Conference 2015 (2015) 7328.pdf AN ESTIMATE OF THE ELEMENTAL COMPOSITION OF LUCA. Aditya Chopra1 and Charles H. Lineweaver1, 1Planetary Science Institute, Research School of Earth Sciences and Research School of Astronomy and Astrophysics, Australian National University, [email protected], [email protected] A number of genomic and proteomic features of composition of life, we attempt to account for life on Earth, like the 16S ribosomal RNA gene, have differences in composition between species and other been highly conserved over billions of years. Genetic phylogenetic taxa (Fig. 2) by weighting datasets such and proteomic conservation translates to conservation that the result represents the root of prokaryotic life of metabolic pathways across taxa. It follows that the (LUCA). Variations in composition between data sets stoichiometry of the elements that make up some of that can be attributed to different growth stages or the biomolecules will be conserved. By extension, the environmental factors are used as estimates of the elemental make up of the whole organism is a uncertainty associated with the average abundances for relatively conserved feature of life on Earth [1,2]. each taxa. We describe how average bulk elemental Euryarchaeota 1a Methanococci, Methanobacteria, Methanopyri 7 Euryarchaeota 1b abundances in extant life can yield an indirect estimate 6 Thermoplasmata, Methanomicrobia, Halobacteria, Archaeoglobi Euryarchaeota 2 4 Thermococci of relative abundances of elements in the Last Crenarchaeota Sulfolobus, Thermoproteus ? Thaumarchaeota Universal Common Ancestor (LUCA). The results Cenarchaeum ? Korarchaeota ARMAN could give us important hints about the stoichiometry ? 2 Archaeal Richmond Mine Acidophilic Nanoorganisms Nanoarchaeota of the environment where LUCA existed and perhaps Archaea Terrabacteria clues to the processes involved in the origin and early Actinobacteria, Deinococcus-Thermus, 1 Cyanobacteria, Life Chloroflexi, 9 evolution of life [3]. -

Do Ultrastable Proteins from Hyperthermophiles Have High Or Low Conformational Rigidity?
Commentary Do ultrastable proteins from hyperthermophiles have high or low conformational rigidity? Rainer Jaenicke* Institute of Biophysics and Physical Biochemistry, University of Regensburg, D-93040 Regensburg, Germany ife on earth has an unbelievable adaptive parisons of their protein inventories with Lcapacity. Except for centers of volcanic those of suitable mesophilic counterparts, a activity, the entire surface of our planet is a wealth of data has been accumulated that biosphere. In this context, the most surpris- indicated that stabilization involves all levels ing discovery in our lifetime was the expan- of the hierarchy of protein structure, i.e., sion from the anthropocentrically defined secondary, supersecondary, tertiary, and ‘‘normal temperature’’ of mesophiles quaternary interactions. The common con- (Ͻ40°C) to the optimum temperature range clusion from model studies was that the of hyperthermophiles around and above the stability of proteins from extremophiles is boiling point of water. That in this class of optimized to maintain corresponding func- microorganisms high temperature is re- tional states under a given set of environ- quired for growth rather than tolerated im- mental conditions. For the standard state at plies that the whole repertoire of their bi- 25°C, enhanced thermal stability of hyper- omolecules must be sufficiently stable to thermophile proteins would then be the allow the cellular microcosm to work. The result of enhanced conformational rigidity Fig. 1. Three-dimensional structure of rubre- strategies nature has used to stabilize the in their folded native state (5). doxin from P. furiosus. Numbered residues mark inventory of the cell, especially proteins, Evidence from recent amide hydrogen the most slowly exchanging hydrogens, close to under extreme conditions are still enig- exchange experiments reported in this is- the two cysteine knuckles (7–9). -

Bhattacharya.1999.Thermophiles.Pdf
THE PHYLOGENY OF THERMOPHILES AND HYPERTHERMOPHILES AND THE THREE DOMAINS OF LIFE The Phylogeny of Thermophiles DEBASHISH BHATTACHARYA University of Iowa Department of Biological Sciences Biology Building, Iowa City, Iowa 52242-1324 United States THOMAS FRIEDL Department of Biology, General Botany University of Kaiserslautern P.O. Box 3049, D-67653 Kaiserslautern, Germany HEIKO SCHMIDT Deutsches Krebsforschungszentrum Theoretische Bioinformatik Im Neuenheimer Feld 280 , D-69120 Heidelberg, Germany 1. Introduction The nature of the first cells and the environment in which they lived are two of the most interesting problems in evolutionary biology. All living things are descendents of these primordial cells and are divided into three fundamental lineages or domains, Archaea (formerly known as Archaebacteria), Bacteria (formerly known as Eubacteria), and the Eucarya (formerly known as Eukaryotes, Woese et al. 1990). The Archaea and Bacteria are prokaryotic domains whereas the Eucarya includes all other living things that have a nucleus (i.e., the genetic material is separated from the cytoplasm by a nuclear envelope). The observation of the three primary domains, first made on the basis of small subunit (i.e., 16S, 18S) ribosomal DNA (rDNA) sequence comparisons (Woese 1987), has created a framework with which the nature of the last common ancestor (LCA) can be addressed. In this review we present phylogenies of the prokaryotic domains to understand the origin and distribution of the thermophiles (organisms able to grow in temperatures > 45°C) and the hyperthermophiles (organisms able to grow in temperatures > 80°C). Hyperthermophiles are limited to the Archaea and Bacteria. In addition, we inspect the distribution of extremophiles within the cyanobacteria. -

Universidad Politécnica De Madrid Escuela Técnica Superior De Ingeniería Agronómica Alimentaria Y De Biosistemas Departamento De Biotecnología-Biología Vegetal
UNIVERSIDAD POLITÉCNICA DE MADRID ESCUELA TÉCNICA SUPERIOR DE INGENIERÍA AGRONÓMICA ALIMENTARIA Y DE BIOSISTEMAS DEPARTAMENTO DE BIOTECNOLOGÍA-BIOLOGÍA VEGETAL TESIS DOCTORAL Caracterización de una nueva proteína hipertermófila NifB y su papel en el mecanismo de síntesis de NifB-co, precursor del grupo metálico de FeMo-co Author: Alessandro Scandurra Director: Prof. Luis Manuel Rubio Herrero Co-director: Dr. Simon Jean Marius Arragain i ii UNIVERSIDAD POLITÉCNICA DE MADRID ESCUELA TÉCNICA SUPERIOR DE INGENIERÍA AGRONÓMICA ALIMENTARIA Y DE BIOSISTEMAS DEPARTAMENTO DE BIOTECNOLOGÍA-BIOLOGÍA VEGETAL Memoria presentada por D. Alessandro Scandurra para optar al grado de Doctor Director Dr. Luis Manuel Rubio Herrero Profesor Titular UPM Madrid, 2017 © This copy oF the thesis has been supplied on condition that anyone who consults it is understood to recognize that its copyright rest with the author and that no quotation From the thesis, or any inFormation derived therefrom may be published without the author’s prior, written consent. I II AcKnowledgments As each journey, also this one has an end (Thanks goodness). It was a long trip, with several changes oF direction. Like For many other people, the PhD adventure was not easy, but I have to admit that I was lucky enough to meet really unique people, that became special Friendships. First oF all, I want to thank my Family, especially my mother and my Father, who always push me to go Forward in my life and to Follow my dreams, ready to pay the sacrifice oF the distance. This expirience has helped me to understand how easy is to be a son and how challenging is to be a parent.