Blueprint Genetics Brachydactyly / Syndactyly Panel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Novel Locus for Brachydactyly Type A1 on Chromosome 5P13.3-P13.2 C M Armour, M E Mccready, a Baig,Agwhunter, D E Bulman
186 LETTERS TO JMG J Med Genet: first published as 10.1136/jmg.39.3.189 on 1 March 2002. Downloaded from A novel locus for brachydactyly type A1 on chromosome 5p13.3-p13.2 C M Armour, M E McCready, A Baig,AGWHunter, D E Bulman ............................................................................................................................. J Med Genet 2002;39:186–189 he brachydactylies are a group of inherited disorders METHODS characterised by shortened or malformed digits that are The linkage study comprised 34 members including 20 Tthought to be the result of abnormal growth of the affected subjects and was conducted after approval by the phalanges and/or metacarpals. First classified by Bell into Children’s Hospital of Eastern Ontario Ethics Review Com- types A, B, C, D, and E, they were reclassified by Temtamy and mittee. McKusick1 and Fitch.2 Brachydactyly type A1 (BDA1, MIM Peripheral blood samples were taken with informed 112500) is characterised by shortened or absent middle consent from all participating family members, and a stand- phalanges. Often the second and fifth digits, as well as the first ard protocol was used to isolate DNA. A genome wide scan proximal phalanx, are the most severely affected. In addition, was initiated using 36 primer sets from the MAPPAIRS™ all of the small tubular bones tend to be reduced in size and microsatellite markers (Research Genetics, Huntsville, Ala- the metacarpals may be shortened, particularly the fifth bama), encompassing markers from 16 chromosomes. metacarpal. Radial/ulnar clinodactyly, as well as malformed or Particular emphasis was placed on markers from chromo- 11 absent epiphyses, have also been reported.12 Complex some 5 and 17, based on the report by Fukushima et al syndromes have been described in which BDA1 is one of a describing a translocation between 5q11.2 and 17q23 in a girl number of manifestations.3 with Klippel-Feil anomaly and BDA1. -

Frontosphenoidal Synostosis: a Rare Cause of Unilateral Anterior Plagiocephaly
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by RERO DOC Digital Library Childs Nerv Syst (2007) 23:1431–1438 DOI 10.1007/s00381-007-0469-4 ORIGINAL PAPER Frontosphenoidal synostosis: a rare cause of unilateral anterior plagiocephaly Sandrine de Ribaupierre & Alain Czorny & Brigitte Pittet & Bertrand Jacques & Benedict Rilliet Received: 30 March 2007 /Published online: 22 September 2007 # Springer-Verlag 2007 Abstract Conclusion Frontosphenoidal synostosis must be searched Introduction When a child walks in the clinic with a in the absence of a coronal synostosis in a child with unilateral frontal flattening, it is usually associated in our anterior unilateral plagiocephaly, and treated surgically. minds with unilateral coronal synostosis. While the latter might be the most common cause of anterior plagiocephaly, Keywords Craniosynostosis . Pediatric neurosurgery. it is not the only one. A patent coronal suture will force us Anterior plagiocephaly to consider other etiologies, such as deformational plagio- cephaly, or synostosis of another suture. To understand the mechanisms underlying this malformation, the development Introduction and growth of the skull base must be considered. Materials and methods There have been few reports in the Harmonious cranial growth is dependent on patent sutures, literature of isolated frontosphenoidal suture fusion, and and any craniosynostosis might lead to an asymmetrical we would like to report a series of five cases, as the shape of the skull. The anterior skull base is formed of recognition of this entity is important for its treatment. different bones, connected by sutures, fusing at different ages. The frontosphenoidal suture extends from the end of Presented at the Consensus Conference on Pediatric Neurosurgery, the frontoparietal suture, anteriorly and inferiorly in the Rome, 1–2 December 2006. -

The Genetic Heterogeneity of Brachydactyly Type A1: Identifying the Molecular Pathways
The genetic heterogeneity of brachydactyly type A1: Identifying the molecular pathways Lemuel Jean Racacho Thesis submitted to the Faculty of Graduate Studies and Postdoctoral Studies in partial fulfillment of the requirements for the Doctorate in Philosophy degree in Biochemistry Specialization in Human and Molecular Genetics Department of Biochemistry, Microbiology and Immunology Faculty of Medicine University of Ottawa © Lemuel Jean Racacho, Ottawa, Canada, 2015 Abstract Brachydactyly type A1 (BDA1) is a rare autosomal dominant trait characterized by the shortening of the middle phalanges of digits 2-5 and of the proximal phalange of digit 1 in both hands and feet. Many of the brachymesophalangies including BDA1 have been associated with genetic perturbations along the BMP-SMAD signaling pathway. The goal of this thesis is to identify the molecular pathways that are associated with the BDA1 phenotype through the genetic assessment of BDA1-affected families. We identified four missense mutations that are clustered with other reported BDA1 mutations in the central region of the N-terminal signaling peptide of IHH. We also identified a missense mutation in GDF5 cosegregating with a semi-dominant form of BDA1. In two families we reported two novel BDA1-associated sequence variants in BMPR1B, the gene which codes for the receptor of GDF5. In 2002, we reported a BDA1 trait linked to chromosome 5p13.3 in a Canadian kindred (BDA1B; MIM %607004) but we did not discover a BDA1-causal variant in any of the protein coding genes within the 2.8 Mb critical region. To provide a higher sensitivity of detection, we performed a targeted enrichment of the BDA1B locus followed by high-throughput sequencing. -

Genetics of Congenital Hand Anomalies
G. C. Schwabe1 S. Mundlos2 Genetics of Congenital Hand Anomalies Die Genetik angeborener Handfehlbildungen Original Article Abstract Zusammenfassung Congenital limb malformations exhibit a wide spectrum of phe- Angeborene Handfehlbildungen sind durch ein breites Spektrum notypic manifestations and may occur as an isolated malforma- an phänotypischen Manifestationen gekennzeichnet. Sie treten tion and as part of a syndrome. They are individually rare, but als isolierte Malformation oder als Teil verschiedener Syndrome due to their overall frequency and severity they are of clinical auf. Die einzelnen Formen kongenitaler Handfehlbildungen sind relevance. In recent years, increasing knowledge of the molecu- selten, besitzen aber aufgrund ihrer Häufigkeit insgesamt und lar basis of embryonic development has significantly enhanced der hohen Belastung für Betroffene erhebliche klinische Rele- our understanding of congenital limb malformations. In addi- vanz. Die fortschreitende Erkenntnis über die molekularen Me- tion, genetic studies have revealed the molecular basis of an in- chanismen der Embryonalentwicklung haben in den letzten Jah- creasing number of conditions with primary or secondary limb ren wesentlich dazu beigetragen, die genetischen Ursachen kon- involvement. The molecular findings have led to a regrouping of genitaler Malformationen besser zu verstehen. Der hohe Grad an malformations in genetic terms. However, the establishment of phänotypischer Variabilität kongenitaler Handfehlbildungen er- precise genotype-phenotype correlations for limb malforma- schwert jedoch eine Etablierung präziser Genotyp-Phänotyp- tions is difficult due to the high degree of phenotypic variability. Korrelationen. In diesem Übersichtsartikel präsentieren wir das We present an overview of congenital limb malformations based Spektrum kongenitaler Malformationen, basierend auf einer ent- 85 on an anatomic and genetic concept reflecting recent molecular wicklungsbiologischen, anatomischen und genetischen Klassifi- and developmental insights. -

Syndromic Ear Anomalies and Renal Ultrasounds
Syndromic Ear Anomalies and Renal Ultrasounds Raymond Y. Wang, MD*; Dawn L. Earl, RN, CPNP‡; Robert O. Ruder, MD§; and John M. Graham, Jr, MD, ScD‡ ABSTRACT. Objective. Although many pediatricians cific MCA syndromes that have high incidences of renal pursue renal ultrasonography when patients are noted to anomalies. These include CHARGE association, Townes- have external ear malformations, there is much confusion Brocks syndrome, branchio-oto-renal syndrome, Nager over which specific ear malformations do and do not syndrome, Miller syndrome, and diabetic embryopathy. require imaging. The objective of this study was to de- Patients with auricular anomalies should be assessed lineate characteristics of a child with external ear malfor- carefully for accompanying dysmorphic features, includ- mations that suggest a greater risk of renal anomalies. We ing facial asymmetry; colobomas of the lid, iris, and highlight several multiple congenital anomaly (MCA) retina; choanal atresia; jaw hypoplasia; branchial cysts or syndromes that should be considered in a patient who sinuses; cardiac murmurs; distal limb anomalies; and has both ear and renal anomalies. imperforate or anteriorly placed anus. If any of these Methods. Charts of patients who had ear anomalies features are present, then a renal ultrasound is useful not and were seen for clinical genetics evaluations between only in discovering renal anomalies but also in the diag- 1981 and 2000 at Cedars-Sinai Medical Center in Los nosis and management of MCA syndromes themselves. Angeles and Dartmouth-Hitchcock Medical Center in A renal ultrasound should be performed in patients with New Hampshire were reviewed retrospectively. Only pa- isolated preauricular pits, cup ears, or any other ear tients who underwent renal ultrasound were included in anomaly accompanied by 1 or more of the following: the chart review. -

Osteodystrophy-Like Syndrome Localized to 2Q37
Am. J. Hum. Genet. 56:400-407, 1995 Brachydactyly and Mental Retardation: An Albright Hereditary Osteodystrophy-like Syndrome Localized to 2q37 L. C. Wilson,"7 K. Leverton,3 M. E. M. Oude Luttikhuis,' C. A. Oley,4 J. Flint,5 J. Wolstenholme,4 D. P. Duckett,2 M. A. Barrow,2 J. V. Leonard,6 A. P. Read,3 and R. C. Trembath' 'Departments of Genetics and Medicine, University of Leicester, and 2Leicestershire Genetics Centre, Leicester Royal Infirmary, Leicester, 3Department of Medical Genetics, St Mary's Hospital, Manchester. 4Department of Human Genetics, University of Newcastle upon Tyne, Newcastle upon Tyne; 5MRC Molecular Haematology Unit, Institute of Molecular Medicine, Oxford; and 6Medical Unit and 7Mothercare Unit for Clinical Genetics and Fetal Medicine, Institute of Child Health, London Summary The physical feature brachymetaphalangia refers to shortening of either the metacarpals and phalanges of the We report five patients with a combination ofbrachymeta- hands or of the equivalent bones in the feet. The combina- phalangia and mental retardation, similar to that observed tion of brachymetaphalangia and mental retardation occurs in Albright hereditary osteodystrophy (AHO). Four pa- in Albright hereditary osteodystrophy (AHO) (Albright et tients had cytogenetically visible de novo deletions of chro- al. 1942), a dysmorphic syndrome that is also associated mosome 2q37. The fifth patient was cytogenetically nor- with cutaneous ossification (in si60% of cases [reviewed by mal and had normal bioactivity of the a subunit of Gs Fitch 1982]); round face; and short, stocky build. Affected (Gsa), the protein that is defective in AHO. In this patient, individuals with AHO may have either pseudohypopara- we have used a combination of highly polymorphic molec- thyroidism (PHP), with end organ resistance to parathyroid ular markers and FISH to demonstrate a microdeletion at hormone (PTH) and certain other cAMP-dependent hor- 2q37. -
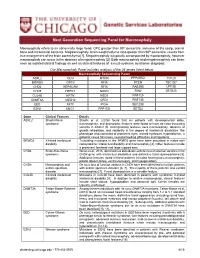
Macrocephaly Information Sheet 6-13-19
Next Generation Sequencing Panel for Macrocephaly Clinical Features: Macrocephaly refers to an abnormally large head, OFC greater than 98th percentile, inclusive of the scalp, cranial bone and intracranial contents. Megalencephaly, brain weight/volume ratio greater than 98th percentile, results from true enlargement of the brain parenchyma [1]. Megalencephaly is typically accompanied by macrocephaly, however macrocephaly can occur in the absence of megalencephaly [2]. Both macrocephaly and megalencephaly can been seen as isolated clinical findings as well as clinical features of a mutli-systemic syndromic diagnosis. Our Macrocephaly Panel includes analysis of the 36 genes listed below. Macrocephaly Sequencing Panel ASXL2 GLI3 MTOR PPP2R5D TCF20 BRWD3 GPC3 NFIA PTEN TBC1D7 CHD4 HEPACAM NFIX RAB39B UPF3B CHD8 HERC1 NONO RIN2 ZBTB20 CUL4B KPTN NSD1 RNF125 DNMT3A MED12 OFD1 RNF135 EED MITF PIGA SEC23B EZH2 MLC1 PPP1CB SETD2 Gene Clinical Features Details ASXL2 Shashi-Pena Shashi et al. (2016) found that six patients with developmental delay, syndrome macrocephaly, and dysmorphic features were found to have de novo truncating variants in ASXL2 [3]. Distinguishing features were macrocephaly, absence of growth retardation, and variability in the degree of intellectual disabilities The phenotype also consisted of prominent eyes, arched eyebrows, hypertelorism, a glabellar nevus flammeus, neonatal feeding difficulties and hypotonia. BRWD3 X-linked intellectual Truncating mutations in the BRWD3 gene have been described in males with disability nonsyndromic intellectual disability and macrocephaly [4]. Other features include a prominent forehead and large cupped ears. CHD4 Sifrim-Hitz-Weiss Weiss et al., 2016, identified five individuals with de novo missense variants in the syndrome CHD4 gene with intellectual disabilities and distinctive facial dysmorphisms [5]. -

Orphanet Journal of Rare Diseases Biomed Central
Orphanet Journal of Rare Diseases BioMed Central Review Open Access Brachydactyly Samia A Temtamy* and Mona S Aglan Address: Department of Clinical Genetics, Human Genetics and Genome Research Division, National Research Centre (NRC), El-Buhouth St., Dokki, 12311, Cairo, Egypt Email: Samia A Temtamy* - [email protected]; Mona S Aglan - [email protected] * Corresponding author Published: 13 June 2008 Received: 4 April 2008 Accepted: 13 June 2008 Orphanet Journal of Rare Diseases 2008, 3:15 doi:10.1186/1750-1172-3-15 This article is available from: http://www.ojrd.com/content/3/1/15 © 2008 Temtamy and Aglan; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Brachydactyly ("short digits") is a general term that refers to disproportionately short fingers and toes, and forms part of the group of limb malformations characterized by bone dysostosis. The various types of isolated brachydactyly are rare, except for types A3 and D. Brachydactyly can occur either as an isolated malformation or as a part of a complex malformation syndrome. To date, many different forms of brachydactyly have been identified. Some forms also result in short stature. In isolated brachydactyly, subtle changes elsewhere may be present. Brachydactyly may also be accompanied by other hand malformations, such as syndactyly, polydactyly, reduction defects, or symphalangism. For the majority of isolated brachydactylies and some syndromic forms of brachydactyly, the causative gene defect has been identified. -

Massachusetts Birth Defects 2002-2003
Massachusetts Birth Defects 2002-2003 Massachusetts Birth Defects Monitoring Program Bureau of Family Health and Nutrition Massachusetts Department of Public Health January 2008 Massachusetts Birth Defects 2002-2003 Deval L. Patrick, Governor Timothy P. Murray, Lieutenant Governor JudyAnn Bigby, MD, Secretary, Executive Office of Health and Human Services John Auerbach, Commissioner, Massachusetts Department of Public Health Sally Fogerty, Director, Bureau of Family Health and Nutrition Marlene Anderka, Director, Massachusetts Center for Birth Defects Research and Prevention Linda Casey, Administrative Director, Massachusetts Center for Birth Defects Research and Prevention Cathleen Higgins, Birth Defects Surveillance Coordinator Massachusetts Department of Public Health 617-624-5510 January 2008 Acknowledgements This report was prepared by the staff of the Massachusetts Center for Birth Defects Research and Prevention (MCBDRP) including: Marlene Anderka, Linda Baptiste, Elizabeth Bingay, Joe Burgio, Linda Casey, Xiangmei Gu, Cathleen Higgins, Angela Lin, Rebecca Lovering, and Na Wang. Data in this report have been collected through the efforts of the field staff of the MCBDRP including: Roberta Aucoin, Dorothy Cichonski, Daniel Sexton, Marie-Noel Westgate and Susan Winship. We would like to acknowledge the following individuals for their time and commitment to supporting our efforts in improving the MCBDRP. Lewis Holmes, MD, Massachusetts General Hospital Carol Louik, ScD, Slone Epidemiology Center, Boston University Allen Mitchell, -

A Review of the Management of Single-Suture Craniosynostosis, Past, Present, and Future
LITERATURE REVIEW J Neurosurg Pediatr 24:622–631, 2019 A review of the management of single-suture craniosynostosis, past, present, and future JNSPG 75th Anniversary Invited Review Article Mark R. Proctor, MD,1 and John G. Meara, MD, DMD, MBA2 Departments of 1Neurosurgery and 2Plastic and Oral Surgery, Boston Children’s Hospital, Boston, Massachusetts BACKGROUND Craniosynostosis is a condition in which 2 or more of the skull bones fuse prematurely. The spectrum of the disorder most commonly involves the closure of a single suture in the skull, but it can also involve syndromic di- agnoses in which multiple skull bones and/or bones outside of the cranium are affected. Craniosynostosis can result in cosmetic deformity as well as potential limitations in brain growth and development, and the neurocognitive impact of the condition is just starting to be studied more thoroughly. Our knowledge regarding the genetics of this condition has also evolved substantially. In this review, the authors explore the medical and surgical advancements in understanding and treating this condition over the past century, with a focus on how the diagnosis and treatment have evolved. METHODS In this review article, the authors, who are the leaders of a craniofacial team at a major academic pediatric hospital, focus on single-suture craniosynostosis (SSC) affecting the 6 major cranial sutures and discuss the evolution of the treatment of SSC from its early history in modern medicine through the current state of the art and future trends. This discussion is based on the authors’ broad experience and a comprehensive review of the literature. SUMMARY The management of SSC has evolved substantially over the past 100 years. -

Classific-Ation and Identification of Inherited Brachydactylies
Journal of Medical Genetics, 1979, 16, 36-44 Classific-ation and identification of inherited brachydactylies NAOMI FITCH From the Lady Davis Institute for Medical Research, Jewish General Hospital, Montreal, Quebec, Canada SUMMARY A search for patterns of malformation in the brachydactylies has resulted in new ways to identify the different types. Type A-I can be characterised by a proportionate reduction of the middle phalanges. Type B is thought to be an amputation-like defect. In type C the fourth middle phalanx is usually the longest, and type E (Riccardi and Holmes, 1974) is characterised by short metacarpals and short distal phalanges. Short stature is usually present in type A-1 and type E brachydactyly (Riccardi and Holmes, 1974) and it may be present in some individuals with brachydactyly C. As short children have short hands, it is possible that in patients with very mild expressions of brachydactyly the cause of the short stature may be overlooked. It is suggested that in every child with propor- tionate short stature the hands should be carefully examined. If the hands are disproportionately short, if any distal creases are missing, if there is a shortening, however mild, of any finger, if any metacarpals are short, then it is important to have x-rays to look for brachydactyly A-1, C, or E. Much information is still needed. It is important in future reports to have skeletal surveys, pattern profile analyses, and to note the height of children with brachydactyly C. Most interesting of all will be when fetal limbs of each type become available for study. -

Blueprint Genetics Craniosynostosis Panel
Craniosynostosis Panel Test code: MA2901 Is a 38 gene panel that includes assessment of non-coding variants. Is ideal for patients with craniosynostosis. About Craniosynostosis Craniosynostosis is defined as the premature fusion of one or more cranial sutures leading to secondary distortion of skull shape. It may result from a primary defect of ossification (primary craniosynostosis) or, more commonly, from a failure of brain growth (secondary craniosynostosis). Premature closure of the sutures (fibrous joints) causes the pressure inside of the head to increase and the skull or facial bones to change from a normal, symmetrical appearance resulting in skull deformities with a variable presentation. Craniosynostosis may occur in an isolated setting or as part of a syndrome with a variety of inheritance patterns and reccurrence risks. Craniosynostosis occurs in 1/2,200 live births. Availability 4 weeks Gene Set Description Genes in the Craniosynostosis Panel and their clinical significance Gene Associated phenotypes Inheritance ClinVar HGMD ALPL Odontohypophosphatasia, Hypophosphatasia perinatal lethal, AD/AR 78 291 infantile, juvenile and adult forms ALX3 Frontonasal dysplasia type 1 AR 8 8 ALX4 Frontonasal dysplasia type 2, Parietal foramina AD/AR 15 24 BMP4 Microphthalmia, syndromic, Orofacial cleft AD 8 39 CDC45 Meier-Gorlin syndrome 7 AR 10 19 EDNRB Hirschsprung disease, ABCD syndrome, Waardenburg syndrome AD/AR 12 66 EFNB1 Craniofrontonasal dysplasia XL 28 116 ERF Craniosynostosis 4 AD 17 16 ESCO2 SC phocomelia syndrome, Roberts syndrome