Genetic Stock Structure of New Zealand Fish and the Use of Genomics in Fisheries Management: an Overview and Outlook
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reef Fish Monitoring Te Tapuwae O Rongokako Marine Reserve
Reef Fish Monitoring Te Tapuwae o Rongokako Marine Reserve Technical Support - Marine East Coast Hawke’s Bay Conservancy Debbie Freeman OCTOBER 2005 Published By Department of Conservation East Coast Hawkes Bay Conservancy PO Box 668 Gisborne 4040, New Zeland Cover: Banded wrasse Photo: I. Nilsson Title page: Koheru Photo: M. Blackwell Acknowledgments: Blue maomao Photo: J. Quirk © Copyright October 2005, New Zealand Department of Conservation ISSN 1175-026X ISBN 978-0-478-14143-6 (paperback) ISBN 978-0-478-14193-1 (Web pdf) Techincal Support Series Number: 25 In the interest of forest conservation, DOC Science Publishing supports paperless electronic publishing. When printing, recycled paper is used wherever possible. C ontents Abstract 4 Introduction 5 Methods 6 Results 10 Discussion 20 Fish fauna 20 Protection effects 21 Reserve age and design 21 Experimental design and monitoring methods 22 Illegal fishing 23 Environmental factors 24 Acknowledgements 24 References 25 Abstract Reef fish monitoring was undertaken within and surrounding Te Tapuwae o Rongokako Marine Reserve, on the North Island’s East Coast, between 2000 and 2004. The objective of the monitoring was to describe the reef fish communities and to establish whether populations within the marine reserve were demonstrating any changes in abundance or size that could be attributable to the removal of fishing pressure. The underwater visual census method was used to survey the four lo- cations (marine reserve and three non-reserve locations). It was found that all four locations were characterised by moderate densities of spot- ties, scarlet wrasse and reef-associated planktivores such as blue maomao, sweep and butterfly perch. -

A Checklist of Fishes of the Aldermen Islands, North-Eastern New Zealand, with Additions to the Fishes of Red Mercury Island
13 A CHECKLIST OF FISHES OF THE ALDERMEN ISLANDS, NORTH-EASTERN NEW ZEALAND, WITH ADDITIONS TO THE FISHES OF RED MERCURY ISLAND by Roger V. Grace* SUMMARY Sixty-five species of marine fishes are listed for the Aldermen Islands, and additions made to an earlier list for Red Mercury Island (Grace, 1972), 35 km to the north. Warm water affinities of the faunas are briefly discussed. INTRODUCTION During recent years, and particularly the last four years, over 30 species of fishes have been added to the New Zealand fish fauna through observation by divers, mainly at the Poor Knights Islands (Russell, 1971; Stephenson, 1970, 1971; Doak, 1972; Whitley, 1968). A high proportion of the fishes of northern New Zealand have strong sub-tropical affinities (Moreland, 1958), and there is considerable evidence (Doak, 1972) to suggest that many of the recently discovered species are new arrivals from tropical and subtropical areas. These fishes probably arrive as eggs or larvae, carried by favourable ocean currents, and find suitable habitats for their development at the Poor Knights Islands, where the warm currents that transported the young fish or eggs maintain a water temperature higher than that on the adjacent coast, or islands to the south. Unless these fishes are able to establish breeding populations in New Zealand waters, they are likely to be merely transient. If they become established, they may begin to spread and colonise other off-shore islands and the coast. In order to monitor any spreading of new arrivals, or die-off due to inability to breed, it is desirable to compile a series of fish lists, as complete as possible, for the off-shore islands of the north-east coast of New Zealand. -

Latris Lineata) in a Data Limited Situation
Assessing the population dynamics and stock viability of striped trumpeter (Latris lineata) in a data limited situation Sean Tracey B. App. Sci. [Fisheries](AMC) A thesis submitted for the degree of Doctor of Philosophy University of Tasmania February 2007 Supervisors Dr. J. Lyle Dr. A. Hobday For my family...Anj and Kails Statement of access I, the undersigned, the author of this thesis, understand that the University of Tas- mania will make it available for use within the university library and, by microfilm or other photographic means, and allow access to users in other approved libraries. All users consulting this thesis will have to sign the following statement: ‘In consulting this thesis I agree not to copy or closely paraphrase it in whole or in part, or use the results in any other work (written or otherwise) without the signed consent of the author; and to make proper written acknowledgment for any other assistance which I have obtained from it.’ Beyond this, I do not wish to place any restrictions on access to this thesis. Signed: .......................................Date:........................................ Sean Tracey Candidate University of Tasmania Declaration I declare that this thesis is my own work and has not been submitted in any form for another degree or diploma at any university or other institution of tertiary edu- cation. Information derived from the published or unpublished work of others has been acknowledged in the text and a list of references is given. Signed: .......................................Date:........................................ Sean Tracey Candidate University of Tasmania Statement of co-authorship Chapters 2 – 5 of this thesis have been prepared as scientific manuscripts. -

Congolli (Pseudaphritis Urvillii) and Australian Salmon (Arripis Truttaceus and A
Inland Waters and Catchment Ecology Diet and trophic characteristics of mulloway (Argyrosomus japonicus), congolli (Pseudaphritis urvillii) and Australian salmon (Arripis truttaceus and A. trutta) in the Coorong George Giatas and Qifeng Ye SARDI Publication No. F2015/000479-1 SARDI Research Report Series No. 858 SARDI Aquatics Sciences PO Box 120 Henley Beach SA 5022 September 2015 Giatas and Ye (2015) Diet of three fish species in the Coorong Diet and trophic characteristics of mulloway (Argyrosomus japonicus), congolli (Pseudaphritis urvillii) and Australian salmon (Arripis truttaceus and A. trutta) in the Coorong George Giatas and Qifeng Ye SARDI Publication No. F2015/000479-1 SARDI Research Report Series No. 858 September 2015 II Giatas and Ye (2015) Diet of three fish species in the Coorong This publication may be cited as: Giatas, G.C. and Ye, Q. (2015). Diet and trophic characteristics of mulloway (Argyrosomus japonicus), congolli (Pseudaphritis urvillii) and Australian salmon (Arripis truttaceus and A. trutta) in the Coorong. South Australian Research and Development Institute (Aquatic Sciences), Adelaide. SARDI Publication No. F2015/000479-1. SARDI Research Report Series No. 858. 81pp. South Australian Research and Development Institute SARDI Aquatic Sciences 2 Hamra Avenue West Beach SA 5024 Telephone: (08) 8207 5400 Facsimile: (08) 8207 5406 http://www.pir.sa.gov.au/research DISCLAIMER The authors warrant that they have taken all reasonable care in producing this report. The report has been through the SARDI internal review process, and has been formally approved for release by the Research Chief, Aquatic Sciences. Although all reasonable efforts have been made to ensure quality, SARDI does not warrant that the information in this report is free from errors or omissions. -

Jeremy Lyle Curriculum Vitae
JEREMY LYLE Biography Dr Jeremy Lyle is a Senior Research Fellow at the Institute for Marine and Antarctic Studies. He is interested in the field of fisheries ecology and biology with a particular focus on understanding fish population dynamics, the impacts of fishing on fish stocks and the characteristics of the recreational as well as commercial fishing sectors. After completing his PhD at the University of Liverpool (UK), Jeremy was employed as a fisheries scientist in the Northern Territory where he led a major initiative to develop a shark fishery off northern Australia. From there he joined the then Tasmanian Sea Fisheries Department and later the University of Tasmania. He has worked on a wide variety of commercial fisheries, including offshore trawl, small pelagics and coastal fisheries, as well as conducting recreational fishing surveys over many years. The primary focus of his research has been to understand the impacts of fishing on target and non-target species and provide the science required to support the sustainable management of the fish stocks. Jeremy's research interests include understanding the impacts of fishing activities on marine resources and how these can be developed and managed sustainably. In relation to recreational fisheries, he has a strong interest in the development of cost-effective survey methods as well as understanding the dynamics and drivers of fishing participation and their implications for resource sharing and management. CURRICULUM VITAE Name: Jeremy Martin Lyle Address: Fisheries and Aquaculture Centre Institute for Marine and Antarctic Studies University of Tasmania Private Bag 49 Hobart TAS 7001 Australia Telephone: Work + 61 3 6227 7255 Mob: +61 407 277 426 Email: [email protected] Current situation: Senior Research Fellow, Fisheries Program Academic History University of Liverpool (Department of Marine Biology), England. -

Catch and Effort Data
Chapter 11: The Inland Waters River Fishery Information System CHAPTER 11: INLAND WATERS RIVER FISHERY INFORMATION SYSTEM 11.1 Background The Inland Waters River fishery is a multi-species, multi-gear fishery encompassing the South Australian sector of the River Murray and its backwaters (Map 11.1). Historically, the fishery was based on harvesting Murray cod (Maccullochella peelii peelii), golden perch (Macquaria ambigua), and bony bream (Nematalosa erebi). The fishery was restructured in July 2003 to a non-native dominate fishery and the commercial fishing for significant native species is now prohibited. The fishery is now based predominantly on the taking European carp (Cyprinus carpio) and redfin perch (Perca fluviatilis). There are a total of 6 licence holders who operate within the fishery. Prior to the restructure 30 license holders fished the river, each with a designated reach. Up to December 2007 SARDI production tables hold in excess of 270,000 daily records. Data is held from 1984/85 to current and is continually being updated. Refer to Figure 11.3 for an entity relationship diagram of the inland waters system that encompasses the River Fishery. South Australian Aquatic Sciences: Information Systems and Database Support Program SARDI Aquatic Sciences Information Systems Quality Assurance and Data Integrity Report (June 2009) 81 Chapter 11: The Inland Waters River Fishery Information System Map 11.1: The inland waters river fishery – River Murray area designations. South Australian Aquatic Sciences: Information Systems and Database Support Program SARDI Aquatic Sciences Information Systems Quality Assurance and Data Integrity Report (June 2009) 82 Chapter 11: The Inland Waters River Fishery Information System 11.2 Research Logbook Information Each licence holder is required to submit a daily fishing return by the 15th day of each month detailing their fishing activities for the previous month (Figure 11.1). -

New Zealand Fishes a Field Guide to Common Species Caught by Bottom, Midwater, and Surface Fishing Cover Photos: Top – Kingfish (Seriola Lalandi), Malcolm Francis
New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing Cover photos: Top – Kingfish (Seriola lalandi), Malcolm Francis. Top left – Snapper (Chrysophrys auratus), Malcolm Francis. Centre – Catch of hoki (Macruronus novaezelandiae), Neil Bagley (NIWA). Bottom left – Jack mackerel (Trachurus sp.), Malcolm Francis. Bottom – Orange roughy (Hoplostethus atlanticus), NIWA. New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing New Zealand Aquatic Environment and Biodiversity Report No: 208 Prepared for Fisheries New Zealand by P. J. McMillan M. P. Francis G. D. James L. J. Paul P. Marriott E. J. Mackay B. A. Wood D. W. Stevens L. H. Griggs S. J. Baird C. D. Roberts‡ A. L. Stewart‡ C. D. Struthers‡ J. E. Robbins NIWA, Private Bag 14901, Wellington 6241 ‡ Museum of New Zealand Te Papa Tongarewa, PO Box 467, Wellington, 6011Wellington ISSN 1176-9440 (print) ISSN 1179-6480 (online) ISBN 978-1-98-859425-5 (print) ISBN 978-1-98-859426-2 (online) 2019 Disclaimer While every effort was made to ensure the information in this publication is accurate, Fisheries New Zealand does not accept any responsibility or liability for error of fact, omission, interpretation or opinion that may be present, nor for the consequences of any decisions based on this information. Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries website at http://www.mpi.govt.nz/news-and-resources/publications/ A higher resolution (larger) PDF of this guide is also available by application to: [email protected] Citation: McMillan, P.J.; Francis, M.P.; James, G.D.; Paul, L.J.; Marriott, P.; Mackay, E.; Wood, B.A.; Stevens, D.W.; Griggs, L.H.; Baird, S.J.; Roberts, C.D.; Stewart, A.L.; Struthers, C.D.; Robbins, J.E. -

Experimental Support Towards a Metabolic Proxy in Fish Using Otolith Carbon Isotopes Jasmin C
© 2020. Published by The Company of Biologists Ltd | Journal of Experimental Biology (2020) 223, jeb217091. doi:10.1242/jeb.217091 RESEARCH ARTICLE Experimental support towards a metabolic proxy in fish using otolith carbon isotopes Jasmin C. Martino1,*,‡, ZoëA. Doubleday1,*, Ming-Tsung Chung2 and Bronwyn M. Gillanders1 ABSTRACT field metabolic rates can be difficult to measure in fish. The doubly Metabolic rate underpins our understanding of how species survive, labelled water method, which involves measuring the elimination of reproduce and interact with their environment, but can be difficult to an introduced oxygen isotope signature, is used for most air-breathing measure in wild fish. Stable carbon isotopes (δ13C) in ear stones animals but is unsuitable for fish (Nelson, 2016; Treberg et al., 2016). (otoliths) of fish may reflect lifetime metabolic signatures but Metabolic rates have therefore been measured using electromyogram experimental validation is required to advance our understanding of telemetry (Cooke et al., 2004; Quintella et al., 2009), heart rate the relationship. To this end, we reared juvenile Australasian snapper monitoring (Clark et al., 2005; Thorarensen et al., 1996) or (Chrysophrys auratus), an iconic fishery species, at different accelerometry (Metcalfe et al., 2016). However, these approaches ‘ ’ temperatures and used intermittent-flow respirometry to calculate only offer short-term snapshots of metabolic rates in live fish. standard metabolic rate (SMR), maximum metabolic rate (MMR) and Records conserved in hard-calcified structures offer a valuable absolute aerobic scope (AAS). Subsequently, we analysed δ13C and alternative to uncovering long-term and retrospective histories. oxygen isotopes (δ18O) in otoliths using isotope-ratio mass Furthermore, biogeochemical techniques can reconstruct histories spectrometry. -

Biogenic Habitats on New Zealand's Continental Shelf. Part II
Biogenic habitats on New Zealand’s continental shelf. Part II: National field survey and analysis New Zealand Aquatic Environment and Biodiversity Report No. 202 E.G. Jones M.A. Morrison N. Davey S. Mills A. Pallentin S. George M. Kelly I. Tuck ISSN 1179-6480 (online) ISBN 978-1-77665-966-1 (online) September 2018 Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries websites at: http://www.mpi.govt.nz/news-and-resources/publications http://fs.fish.govt.nz go to Document library/Research reports © Crown Copyright – Fisheries New Zealand TABLE OF CONTENTS EXECUTIVE SUMMARY 1 1. INTRODUCTION 3 1.1 Overview 3 1.2 Objectives 4 2. METHODS 5 2.1 Selection of locations for sampling. 5 2.2 Field survey design and data collection approach 6 2.3 Onboard data collection 7 2.4 Selection of core areas for post-voyage processing. 8 Multibeam data processing 8 DTIS imagery analysis 10 Reference libraries 10 Still image analysis 10 Video analysis 11 Identification of biological samples 11 Sediment analysis 11 Grain-size analysis 11 Total organic matter 12 Calcium carbonate content 12 2.5 Data Analysis of Core Areas 12 Benthic community characterization of core areas 12 Relating benthic community data to environmental variables 13 Fish community analysis from DTIS video counts 14 2.6 Synopsis Section 15 3. RESULTS 17 3.1 -

Uva-DARE (Digital Academic Repository)
UvA-DARE (Digital Academic Repository) The adequacy of aging techniques in vertebrates for rapid estimation of population mortality rates from age distributions Zhao, M.; Klaassen, C.A.J.; Lisovski, S.; Klaassen, M. DOI 10.1002/ece3.4854 Publication date 2019 Document Version Other version Published in Ecology and Evolution License CC BY Link to publication Citation for published version (APA): Zhao, M., Klaassen, C. A. J., Lisovski, S., & Klaassen, M. (2019). The adequacy of aging techniques in vertebrates for rapid estimation of population mortality rates from age distributions. Ecology and Evolution, 9(3), 1394-1402. https://doi.org/10.1002/ece3.4854 General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:01 Oct 2021 1 Appendix S4. -
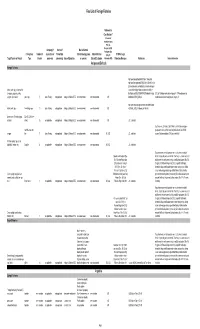
2018 Final LOFF W/ Ref and Detailed Info
Final List of Foreign Fisheries Rationale for Classification ** (Presence of mortality or injury (P/A), Co- Occurrence (C/O), Company (if Source of Marine Mammal Analogous Gear Fishery/Gear Number of aquaculture or Product (for Interactions (by group Marine Mammal (A/G), No RFMO or Legal Target Species or Product Type Vessels processor) processing) Area of Operation or species) Bycatch Estimates Information (N/I)) Protection Measures References Detailed Information Antigua and Barbuda Exempt Fisheries http://www.fao.org/fi/oldsite/FCP/en/ATG/body.htm http://www.fao.org/docrep/006/y5402e/y5402e06.htm,ht tp://www.tradeboss.com/default.cgi/action/viewcompan lobster, rock, spiny, demersal fish ies/searchterm/spiny+lobster/searchtermcondition/1/ , (snappers, groupers, grunts, ftp://ftp.fao.org/fi/DOCUMENT/IPOAS/national/Antigua U.S. LoF Caribbean spiny lobster trap/ pot >197 None documented, surgeonfish), flounder pots, traps 74 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G AndBarbuda/NPOA_IUU.pdf Caribbean mixed species trap/pot are category III http://www.nmfs.noaa.gov/pr/interactions/fisheries/tabl lobster, rock, spiny free diving, loops 19 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G e2/Atlantic_GOM_Caribbean_shellfish.html Queen conch (Strombus gigas), Dive (SCUBA & free molluscs diving) 25 not applicable not applicable Antigua & Barbuda EEZ none documented none documented A/G U.S. trade data Southeastern U.S. Atlantic, Gulf of Mexico, and Caribbean snapper- handline, hook and grouper and other reef fish bottom longline/hook-and-line/ >5,000 snapper line 71 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented N/I, A/G U.S. -

Elephant Fish
Best Fish Guide 2009-2010 How sustainable is New Zealand seafood? (Ecological Assessments) Produced and Published by Royal Forest and Bird Protection Society of New Zealand, Inc. PO Box 631, Level One, 90 Ghuznee Street, Wellington. www.forestandbird.org.nz November 2009 Acknowledgements Forest & Bird with to thank anonymous reviewers for their peer review comments on this draft. We also thank Peta Methias, Annabel Langbein, Martin Bosely, Margaret Brooker, Lois Daish, Kelder Haines, Dobie Blaze, Rohan Horner and Ray McVinnie for permission to use their recipes on the website. Special thanks to our Best Fish Guide Ambassador Dobie Blaze, keyboard player with Fat Freddy’s Drop. Editing: Kirstie Knowles, Barry Weeber and Helen Bain Technical Advisor: Barry Weeber Cover Design: Rob Deliver Cover fish (Tarakihi): Malcolm Francis Photography: Malcolm Francis: blue cod, blue moki, blue shark, butterfish, groper/hapuku, hoki, jack mackerel, john dory, kahawai, kingfish, leather jacket, moonfish, paua, porbeagle shark, red gurnard, red snapper, scallop, school shark, sea perch, snapper, spiny dogfish, tarakihi, trevally and trumpeter. Peter Langlands: blue warehou, cockles, elephantfish, frostfish, lookdown dory, oyster, pale ghost shark, queen scallops, red cod, rig/lemonfish, rubyfish and scampi. Ministry of Fisheries: albacore tuna, bigeye tuna, blue mackerel, pacific bluefin tuna, skipjack tuna, southern bluefin tuna and swordfish. John Holdsworth: gemfish, striped marlin and yellowfin tuna. Kirstie Knowles: sand flounder and rock lobster. Department of Conservation: kina and skate. Quentin Bennett: mako shark. Scott Macindoe: garfish. Jim Mikoz: yellow-eyed mullet. Forest & Bird: arrow squid, dark ghost shark, orange roughy, smooth oreo, packhorse lobster, paddle crabs, stargazer and white warehou.