Identification and Developmental Expression Profiling of Putative
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Network Pharmacology and Traditional Chinese Medicine: Devel- Opment of Anti-Diabetic Therapies Zhongxia Lu1, Wenjun Xu1, Xi Chen2, Changyu Li3* and Yitao Chen1*
ISSN: 2377-3634 Lu et al. Int J Diabetes Clin Res 2017, 4:077 DOI: 10.23937/2377-3634/1410077 Volume 4 | Issue 2 International Journal of Open Access Diabetes and Clinical Research REVIEW ARTICLE Network Pharmacology and Traditional Chinese Medicine: Devel- opment of Anti-Diabetic Therapies Zhongxia Lu1, Wenjun Xu1, Xi Chen2, Changyu Li3* and Yitao Chen1* 1College of Life Sciences, Zhejiang Chinese Medical University, Hangzhou, Zhejiang, China 2College of Traditional Chinese Medicine, Beijing University of Chinese Medicine, China Check for 3College of Pharmacy, Zhejiang Chinese Medical University, Hangzhou, Zhejiang, China updates *Corresponding author: Yitao Chen, MD, College of Life Sciences, Zhejiang Chinese Medical University, Hangzhou, Zhejiang, 310053, China, E-mail: [email protected]; Changyu Li, MD, College of Pharmacy, Zhejiang Chinese Medical University, Hangzhou, Zhejiang, 310053, China, E-mail: [email protected] Abstract Research and Development Dilemma for Anti- Diabetes Drugs Partly due to the failure of single-target drugs, diabetes mel- litus, a chronic metabolic disease with complex pathogene- Type 2 Diabetes Mellitus (T2DM), generally agreed sis and long-term medication requirements, is increasing in to be caused by insulin resistance and/or insulin defi- prevalence worldwide and urgently needs multi-component and multi-target treatments. Traditional Chinese herbs are ciency, constitutes almost 95 percent of all diabetes the principal drug of Chinese medicine, which is effective cases [4]. Because of the pathogenesis, the mainstre- against diabetes. However, Chinese herbs’ mechanism of am anti-diabetic drugs are insulin secretagogues (sul- action is difficult to elucidate due to its multiple components phonylureas and meglitinide analogues), insulin sensi- and multi-target effects. -

Supplementary Materials 1
Supplementary materials 1 Table S1 The characteristics of botanical preparations potentially containing alkenylbenzenes on the Chinese market. Botanical Pin Yin Name Form Ingredients Recommendation for daily intake (g) preparations (汉语) Plant food supplements (PFS) Si Ji Kang Mei Yang Xin Yuan -Rou Dou Kou xylooligosaccharide, isomalt, nutmeg (myristica PFS 1 Fu He Tang Pian tablet 4 tablets (1.4 g) fragrans), galangal, cinnamon, chicken gizzards (四季康美养心源-肉豆蔻复合糖片) Ai Si Meng Hui Xiang fennel seed, figs, prunes, dates, apples, St.Johns 2-4 tablets (2.8-5.6 g) PFS 2 Fu He Pian tablet Breed, jamaican ginger root (爱司盟茴香复合片) Zi Ran Mei Xiao Hui Xiaong Jiao Nang foeniculi powder, cinnamomi cortex, papaya PFS 3 capsule concentrated powder, green oat concentrated powder, 3 capsules (1.8 g) (自然美小茴香胶囊) brewer’s yeast, cabbage, monkey head mushroom An Mei Qi Hui Xiang Cao Ben Fu He Pian fennel seed, perilla seed, cassia seed, herbaceous PFS 4 tablet 1-2 tablets (1.4-2.8 g) (安美奇茴香草本复合片) complex papaya enzymes, bromelain enzymes, lactobacillus An Mei Qi Jiao Su Xian Wei Ying Yang Pian acidophilus, apple fiber, lemon plup fiber, fennel PFS 5 tablet seed, cascara sagrada, jamaican ginger root, herbal 2 tablets (2.7 g) (安美奇酵素纤维营养片) support complex (figs, prunes, dates, apples, St. Johns bread) Table S1 (continued) The characteristics of botanical preparations potentially containing alkenylbenzenes on the Chinese market. Pin Yin Name Botanical Form Ingredients Recommendation for daily intake (g) preparations (汉语) Gan Cao Pian glycyrrhiza uralensis, licorice -

Effect of Wine and Vinegar Processing of Rhizoma Corydalis on the Tissue Distribution of Tetrahydropalmatine, Protopine and Dehydrocorydaline in Rats
Michigan Technological University Digital Commons @ Michigan Tech Michigan Tech Publications 1-18-2012 Effect of wine and vinegar processing of Rhizoma Corydalis on the tissue distribution of tetrahydropalmatine, protopine and dehydrocorydaline in rats Zhiying Dou Tianjin University of Traditional Chinese Medicine Kefeng Li Michigan Technological University Ping Wang Tianjin University of Traditional Chinese Medicine Liu Cao Tianjin University of Traditional Chinese Medicine Follow this and additional works at: https://digitalcommons.mtu.edu/michigantech-p Part of the Biology Commons Recommended Citation Dou, Z., Li, K., Wang, P., & Cao, L. (2012). Effect of wine and vinegar processing of Rhizoma Corydalis on the tissue distribution of tetrahydropalmatine, protopine and dehydrocorydaline in rats. Molecules, 17(1), 951-970. http://doi.org/10.3390/molecules17010951 Retrieved from: https://digitalcommons.mtu.edu/michigantech-p/1969 Follow this and additional works at: https://digitalcommons.mtu.edu/michigantech-p Part of the Biology Commons Molecules 2012, 17, 951-970; doi:10.3390/molecules17010951 OPEN ACCESS molecules ISSN 1420-3049 www.mdpi.com/journal/molecules Article Effect of Wine and Vinegar Processing of Rhizoma Corydalis on the Tissue Distribution of Tetrahydropalmatine, Protopine and Dehydrocorydaline in Rats Zhiying Dou 1,*, Kefeng Li 2, Ping Wang 1 and Liu Cao 1 1 College of Chinese Materia Medica, Tianjin University of Traditional Chinese Medicine, Tianjin 300193, China 2 Department of Biological Sciences, Michigan Technological University, Houghton, MI 49931, USA; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel./Fax: +86-22-5959-6235. Received: 29 November 2011; in revised form: 5 January 2012 / Accepted: 9 January 2012 / Published: 18 January 2012 Abstract: Vinegar and wine processing of medicinal plants are two traditional pharmaceutical techniques which have been used for thousands of years in China. -

Expression of Genes Encoding Acetyl-Coa Carboxylase, Biotin Synthase, and Acetyl-Coa Generating Enzymes in Arabidopsis Thaliana Jinshan Ke Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1997 Expression of genes encoding acetyl-CoA carboxylase, biotin synthase, and acetyl-CoA generating enzymes in Arabidopsis thaliana Jinshan Ke Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Botany Commons, and the Molecular Biology Commons Recommended Citation Ke, Jinshan, "Expression of genes encoding acetyl-CoA carboxylase, biotin synthase, and acetyl-CoA generating enzymes in Arabidopsis thaliana " (1997). Retrospective Theses and Dissertations. 11996. https://lib.dr.iastate.edu/rtd/11996 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly fi"om the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter fece, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely aflfect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. -

NRDC: Generally Recognized As Secret
NRDC Report April 2014 Generally Recognized as Secret: Chemicals Added to Food in the United States Tom Neltner, J.D., Maricel Maffini, Ph.D. Natural Resources Defense Council In April 2014, the Natural Resources Defense Council (NRDC) released a report raising concerns about a loophole in the Food Additives Amendment of 1958 for substances designated by food manufacturers as “generally recognized as safe” (GRAS). The report identified 56 companies that appeared to market 275 chemicals for use in food based on undisclosed GRAS safety determinations. For each chemical we identified in this study, we did not find evidence that FDA had cleared them for use in food. The 1958 law exempted from the formal, extended FDA approval process common food ingredients like vinegar and vegetable oil whose use qualifies as GRAS. It may have appeared reasonable at the time, but that exemption has been stretched into a loophole that has swallowed the law. The exemption allows manufacturers to make safety determinations that the uses of their newest chemicals in food are safe without notifying the FDA. The agency’s attempts to limit these undisclosed GRAS determinations by asking industry to voluntarily inform the FDA about their chemicals are insufficient to ensure the safety of our food in today’s global marketplace with a complex food supply. Furthermore, no other developed country in the world has a system like GRAS to provide oversight of food ingredients. In Table 1 and 2 of the report, NRDC identified the 56 companies and the number of chemicals that each company appeared to market as GRAS without FDA clearance. -

REGULATION of METABOLISM by the ONCOPROTEIN C-MYC by Lia
REGULATION OF METABOLISM BY THE ONCOPROTEIN C-MYC by Lia Rae Edmunds Biochemistry, Washington and Jefferson College, 2008 Submitted to the Graduate Faculty of Molecular Genetics and Developmental Biology in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2015 UNIVERSITY OF PITTSBURGH MOLECULAR GENETICS AND DEVELOPMENTAL BIOLOGY This dissertation was presented by Lia Rae Edmunds It was defended on November, 2015 and approved by Eric S. Goetzman, Ph.D., Medical Genetics Robert M. O’Doherty, Ph.D., Endocrinology Bennet Van Houten, Ph.D., Molecular Genetics and Developmental Biology Dissertation Advisor: Edward V. Prochownik, M.D., Ph.D., Pediatric Hematology/Oncology ii Copyright © by Lia Rae Edmunds 2015 iii REGULATION OF METABOLISM BY THE ONCOPROTEIN C-MYC Lia Rae Edmunds, Ph.D. University of Pittsburgh, 2015 c-Myc (hereafter Myc), a transcription factor that regulates a variety of cellular functions including growth and differentiation, is deregulated in many different types of cancers. Myc regulates the Warburg effect and oncogenic biosynthesis, but also many aspects of metabolism, believed to be a pivotal point of transformation. Myc is known to control glycolysis and glutaminolysis but little is known about the interplay between glucose, amino acid, and fatty acid oxidation. We hypothesize Myc integrates glucose, amino acid, and fatty acid utilization for energy, and either loss- or gain-of-function will disrupt metabolic homeostasis. Loss of Myc in rat fibroblasts elicits a severe energy deficit, including diminished acetyl-coA levels, to which they respond by enhancing FAO and lipid uptake and storage. Using an in vivo model, we found murine hepatocytes respond to Myc ablation with a milder phenotype. -
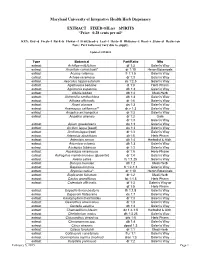
SPIRITS *Price: 0.28 Cents Per Ml*
Maryland University of Integrative Health Herb Dispensary EXTRACT FIXED OIL(s) SPIRITS *Price: 0.28 cents per ml* KEY: Dry=d Fresh=f Bark=b Flower=f Fruit/Seed=s Leaf=l Herb=H Rhizome=z Root=r Stem=st Resin=rsn Note: Part ratio may vary due to supply. Updated 1/29/2015 Type Botanical Part/Ratio Mfg extract Achillea millefolium df 1:3 Galen’s Way extract Aconitum carmichaeli* dr 1:10 Heron Botanicals extract Acorus calamus fr 1:1.5 Galen’s Way extract Actaea racemosa dr 1:3 Galen’s Way extract Aesculus hippocastanum ds 1:2.5 Galen’s Way extract Agathosma betulina dl 1:3 Herb Pharm extract Agrimonia eupatoria dh 1:3 Galen’s Way extract Albizia lebbek db 1:2 Medi-Herb extract Alchemilla xanthochlora dh 1:3 Galen’s Way extract Althaea officinalis dr 1:6 Galen’s Way extract Ammi visnaga ds 1:3 Galen’s Way extract Anemopsis californica** dr,z 1:3 Galen’s Way extract Angelica archangelica dr 1:3 Galen’s Way extract Angelica sinensis dr 1:2 Gaia dr 1:3 Galen’s Way extract Apium graveoloens ds 1:3 Galen’s Way extract Arctium lappa (seed) ds 1:3 Galen’s Way extract Arctium lappa (root) dr 1:3 Galen’s Way extract Artemisia absinthium dh1:5 Herb Pharm extract Artemisia annua dh 1:4 Herbalist & Alch. extract Artemisia vulgaris dh 1:3 Galen’s Way extract Asclepias tuberosa dr 1:3 Galen’s Way extract Asparagus racemosus dr 1:5 Herb-Pharm extract Astragalus membranaceus (glycerite) dr 1:4 Galen’s Way extract Avena sativa fs 1:1.25 Galen’s Way extract Bacopa monnieri dh 1:2 Medi-Herb extract Baptisia tinctoria fr 1:2-1:3 Galen’s Way extract Bryonia cretica* dr 1:10 Heron Botanicals extract Bupleurum falcatum dr 1:2 Medi-Herb extract Cactus grandiflorus fst 1:1.5 Herb Pharm extract Calendula officinalis df 1:3 Galen’s Way or df 1:5 Herb Pharm extract Capsella bursa-pastoris fh 1:1.5 Galen’s Way extract Capsicum frutescens ds 1:7 Galen’s Way extract Caulophyllum thalictroides dr 1:3 Galen’s Way extract Ceanothus americanus dr 1:3 Galen’s Way extract Centella asiatica dh 1:2 Galen’s Way extract Chamaelirium luteum dz 1:4-1:5 Herbalist & Alch. -

(10) Patent No.: US 8119385 B2
US008119385B2 (12) United States Patent (10) Patent No.: US 8,119,385 B2 Mathur et al. (45) Date of Patent: Feb. 21, 2012 (54) NUCLEICACIDS AND PROTEINS AND (52) U.S. Cl. ........................................ 435/212:530/350 METHODS FOR MAKING AND USING THEMI (58) Field of Classification Search ........................ None (75) Inventors: Eric J. Mathur, San Diego, CA (US); See application file for complete search history. Cathy Chang, San Diego, CA (US) (56) References Cited (73) Assignee: BP Corporation North America Inc., Houston, TX (US) OTHER PUBLICATIONS c Mount, Bioinformatics, Cold Spring Harbor Press, Cold Spring Har (*) Notice: Subject to any disclaimer, the term of this bor New York, 2001, pp. 382-393.* patent is extended or adjusted under 35 Spencer et al., “Whole-Genome Sequence Variation among Multiple U.S.C. 154(b) by 689 days. Isolates of Pseudomonas aeruginosa” J. Bacteriol. (2003) 185: 1316 1325. (21) Appl. No.: 11/817,403 Database Sequence GenBank Accession No. BZ569932 Dec. 17. 1-1. 2002. (22) PCT Fled: Mar. 3, 2006 Omiecinski et al., “Epoxide Hydrolase-Polymorphism and role in (86). PCT No.: PCT/US2OO6/OOT642 toxicology” Toxicol. Lett. (2000) 1.12: 365-370. S371 (c)(1), * cited by examiner (2), (4) Date: May 7, 2008 Primary Examiner — James Martinell (87) PCT Pub. No.: WO2006/096527 (74) Attorney, Agent, or Firm — Kalim S. Fuzail PCT Pub. Date: Sep. 14, 2006 (57) ABSTRACT (65) Prior Publication Data The invention provides polypeptides, including enzymes, structural proteins and binding proteins, polynucleotides US 201O/OO11456A1 Jan. 14, 2010 encoding these polypeptides, and methods of making and using these polynucleotides and polypeptides. -

( 12 ) United States Patent
US010208322B2 (12 ) United States Patent ( 10 ) Patent No. : US 10 ,208 , 322 B2 Coelho et al. (45 ) Date of Patent: * Feb . 19, 2019 ( 54 ) IN VIVO AND IN VITRO OLEFIN ( 56 ) References Cited CYCLOPROPANATION CATALYZED BY HEME ENZYMES U . S . PATENT DOCUMENTS 3 , 965 ,204 A 6 / 1976 Lukas et al. (71 ) Applicant: California Institute of Technology , 4 , 243 ,819 A 1 / 1981 Henrick Pasadena , CA (US ) 5 ,296 , 595 A 3 / 1994 Doyle 5 , 703 , 246 A 12 / 1997 Aggarwal et al. 7 , 226 , 768 B2 6 / 2007 Farinas et al. ( 72 ) Inventors : Pedro S . Coelho , Los Angeles, CA 7 , 267 , 949 B2 9 / 2007 Richards et al . (US ) ; Eric M . Brustad , Durham , NC 7 ,625 ,642 B2 12 / 2009 Matsutani et al. (US ) ; Frances H . Arnold , La Canada , 7 ,662 , 969 B2 2 / 2010 Doyle et al. CA (US ) ; Zhan Wang , San Jose , CA 7 ,863 ,030 B2 1 / 2011 Arnold (US ) ; Jared C . Lewis , Chicago , IL 8 ,247 ,430 B2 8 / 2012 Yuan 8 , 993 , 262 B2 * 3 / 2015 Coelho . .. .. .. • * • C12P 7 /62 (US ) 435 / 119 9 ,399 , 762 B26 / 2016 Farwell et al . (73 ) Assignee : California Institute of Technology , 9 , 493 ,799 B2 * 11 /2016 Coelho .. C12P 7162 Pasadena , CA (US ) 2006 / 0030718 AL 2 / 2006 Zhang et al. 2006 / 0111347 A1 5 / 2006 Askew , Jr . et al. 2007 /0276013 AL 11 /2007 Ebbinghaus et al . ( * ) Notice : Subject to any disclaimer , the term of this 2009 /0238790 A2 9 /2009 Sommadosi et al. patent is extended or adjusted under 35 2010 / 0056806 A1 3 / 2010 Warren U . -

Rol De La Vía Autofágica-Lisosomal En La Muerte Celular Inducida Por Manganeso En Células Gliales
Tesis Doctoral Rol de la vía autofágica-lisosomal en la muerte celular inducida por manganeso en células gliales Gorojod, Roxana Mayra 2014-07-17 Este documento forma parte de la colección de tesis doctorales y de maestría de la Biblioteca Central Dr. Luis Federico Leloir, disponible en digital.bl.fcen.uba.ar. Su utilización debe ser acompañada por la cita bibliográfica con reconocimiento de la fuente. This document is part of the doctoral theses collection of the Central Library Dr. Luis Federico Leloir, available in digital.bl.fcen.uba.ar. It should be used accompanied by the corresponding citation acknowledging the source. Cita tipo APA: Gorojod, Roxana Mayra. (2014-07-17). Rol de la vía autofágica-lisosomal en la muerte celular inducida por manganeso en células gliales. Facultad de Ciencias Exactas y Naturales. Universidad de Buenos Aires. Cita tipo Chicago: Gorojod, Roxana Mayra. "Rol de la vía autofágica-lisosomal en la muerte celular inducida por manganeso en células gliales". Facultad de Ciencias Exactas y Naturales. Universidad de Buenos Aires. 2014-07-17. Dirección: Biblioteca Central Dr. Luis F. Leloir, Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires. Contacto: [email protected] Intendente Güiraldes 2160 - C1428EGA - Tel. (++54 +11) 4789-9293 UNIVERSIDAD DE BUENOS AIRES Facultad de Ciencias Exactas y Naturales Departamento de Química Biológica Rol de la vía autofágica- lisosomal en la muerte celular inducida por manganeso en células gliales Tesis doctoral para optar al título de Doctor de la Universidad de Buenos Aires en el área de Química Biológica Lic. Roxana Mayra Gorojod Director de tesis: Dra. -

Proprietary Products Guide
Proprietary Formulas Our proprietary blends are unlike any on the market and offer acute relief as well as tonic support for constitutional health. Highly effective and accessibly priced, our remedies inspire repeat purchases and loyal following. Our practitioner partners love having these on hand, and find they are well matched for many common health concerns. All our formulas are available for distribution under your label and are also offered bulk. Commitment to Quality Superior products are only possible with superior medicinals. We source from the most reputable herb suppliers, including local farms and responsible wildcrafters. We are proud to work only with Chinese herb distributors who employ rigid standards and quality assurance testing, and continually offer us better access to organically grown Chinese medicinals. All of the Western herbs we buy are certified organic, locally grown, or ethically wildcrafted. Each of our vendors goes through our GMP supplier qualification program, and each herb is analyzed against our rigorous specifications to ensure the identity, purity, and potency of every medicinal. We are registered to produce certified organic products by CCOF. Our Approach Launched by a professional herbalist / acupuncturist, and a medical doctor, Five Flavors Herbs bridges the therapeutic traditions of East and West. We combine scientific research and clinical practice with millenniums old theory of healing, offering reliable tools for practitioners and purveyors. Truly unique formulations, our extract line stands out both in approach and effectiveness. Nervous System Remedies Elation (Xiao Yao San +) ~ mood support Lifts the mood and promotes a healthy emotional response to stress and monthly hormonal changes.* Take 15-60 drops in ¼ cup water 3-5 times daily or as directed. -

Bifunctional CYP81AA Proteins Catalyse Identical Hydroxylations but Alternative Regioselective Phenol Couplings in Plant Xanthone Biosynthesis
ARTICLE Received 27 Oct 2015 | Accepted 30 Mar 2016 | Published 5 May 2016 DOI: 10.1038/ncomms11472 OPEN Bifunctional CYP81AA proteins catalyse identical hydroxylations but alternative regioselective phenol couplings in plant xanthone biosynthesis Islam El-Awaad1,2,w, Marco Bocola3, Till Beuerle1,2, Benye Liu1,2 & Ludger Beerhues1,2 Xanthones are natural products present in plants and microorganisms. In plants, their biosynthesis starts with regioselective cyclization of 2,30,4,6-tetrahydroxybenzophenone to either 1,3,5- or 1,3,7-trihydroxyxanthones, catalysed by cytochrome P450 (CYP) enzymes. Here we isolate and express CYP81AA-coding sequences from Hypericum calycinum and H. perforatum in yeast. Microsomes catalyse two consecutive reactions, that is, 30-hydro- xylation of 2,4,6-trihydroxybenzophenone and C–O phenol coupling of the resulting 2,30, 4,6-tetrahydroxybenzophenone. Relative to the inserted 30-hydroxyl, the orthologues Hc/HpCYP81AA1 cyclize via the para position to form 1,3,7-trihydroxyxanthone, whereas the paralogue HpCYP81AA2 directs cyclization to the ortho position, yielding the isomeric 1,3,5- trihydroxyxanthone. Homology modelling and reciprocal mutagenesis reveal the impact of S375, L378 and A483 on controlling the regioselectivity of HpCYP81AA2, which is converted into HpCYP81AA1 by sextuple mutation. However, the reciprocal mutations in HpCYP81AA1 barely affect its regiospecificity. Product docking rationalizes the alternative C–O phenol coupling reactions. Our results help understand the machinery of bifunctional CYPs. 1 Institute of Pharmaceutical Biology, Technische Universita¨t Braunschweig, Mendelssohnstrae 1, Braunschweig 38106, Germany. 2 Center of Pharmaceutical Engineering (PVZ), Technische Universita¨t Braunschweig, Franz-Liszt-Strae 35A, Braunschweig 38106, Germany. 3 Institute of Biotechnology, RWTH Aachen University, Worringerweg 1, Aachen 52074, Germany.