DNA Barcode-Based Assessment of Arthropod Diversity in Canada's
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Chironomidae Hirschkopf
Literatur Chironomidae Gesäuse U.A. zur Bestimmung und Ermittlung der Autökologie herangezogene Literatur: Albu, P. (1972): Două specii de Chironomide noi pentru ştiinţă în masivul Retezat.- St. şi Cerc. Biol., Seria Zoologie, 24: 15-20. Andersen, T.; Mendes, H.F. (2002): Neotropical and Mexican Mesosmittia Brundin, with the description of four new species (Insecta, Diptera, Chironomidae).- Spixiana, 25(2): 141-155. Andersen, T.; Sæther, O.A. (1993): Lerheimia, a new genus of Orthocladiinae from Africa (Diptera: Chironomidae).- Spixiana, 16: 105-112. Andersen, T.; Sæther, O.A.; Mendes, H.F. (2010): Neotropical Allocladius Kieffer, 1913 and Pseudosmittia Edwards, 1932 (Diptera: Chironomidae).- Zootaxa, 2472: 1-77. Baranov, V.A. (2011): New and rare species of Orthocladiinae (Diptera, Chironomidae) from the Crimea, Ukraine.- Vestnik zoologii, 45(5): 405-410. Boggero, A.; Zaupa, S.; Rossaro, B. (2014): Pseudosmittia fabioi sp. n., a new species from Sardinia (Diptera: Chironomidae, Orthocladiinae).- Journal of Entomological and Acarological Research, [S.l.],46(1): 1-5. Brundin, L. (1947): Zur Kenntnis der schwedischen Chironomiden.- Arkiv för Zoologi, 39 A(3): 1- 95. Brundin, L. (1956): Zur Systematik der Orthocladiinae (Dipt. Chironomidae).- Rep. Inst. Freshwat. Drottningholm 37: 5-185. Casas, J.J.; Laville, H. (1990): Micropsectra seguyi, n. sp. du groupe attenuata Reiss (Diptera: Chironomidae) de la Sierra Nevada (Espagne).- Annls Soc. ent. Fr. (N.S.), 26(3): 421-425. Caspers, N. (1983): Chironomiden-Emergenz zweier Lunzer Bäche, 1972.- Arch. Hydrobiol. Suppl. 65: 484-549. Caspers, N. (1987): Chaetocladius insolitus sp. n. (Diptera: Chironomidae) from Lunz, Austria. In: Saether, O.A. (Ed.): A conspectus of contemporary studies in Chironomidae (Diptera). -

Checklist of the Family Chironomidae (Diptera) of Finland
A peer-reviewed open-access journal ZooKeys 441: 63–90 (2014)Checklist of the family Chironomidae (Diptera) of Finland 63 doi: 10.3897/zookeys.441.7461 CHECKLIST www.zookeys.org Launched to accelerate biodiversity research Checklist of the family Chironomidae (Diptera) of Finland Lauri Paasivirta1 1 Ruuhikoskenkatu 17 B 5, FI-24240 Salo, Finland Corresponding author: Lauri Paasivirta ([email protected]) Academic editor: J. Kahanpää | Received 10 March 2014 | Accepted 26 August 2014 | Published 19 September 2014 http://zoobank.org/F3343ED1-AE2C-43B4-9BA1-029B5EC32763 Citation: Paasivirta L (2014) Checklist of the family Chironomidae (Diptera) of Finland. In: Kahanpää J, Salmela J (Eds) Checklist of the Diptera of Finland. ZooKeys 441: 63–90. doi: 10.3897/zookeys.441.7461 Abstract A checklist of the family Chironomidae (Diptera) recorded from Finland is presented. Keywords Finland, Chironomidae, species list, biodiversity, faunistics Introduction There are supposedly at least 15 000 species of chironomid midges in the world (Armitage et al. 1995, but see Pape et al. 2011) making it the largest family among the aquatic insects. The European chironomid fauna consists of 1262 species (Sæther and Spies 2013). In Finland, 780 species can be found, of which 37 are still undescribed (Paasivirta 2012). The species checklist written by B. Lindeberg on 23.10.1979 (Hackman 1980) included 409 chironomid species. Twenty of those species have been removed from the checklist due to various reasons. The total number of species increased in the 1980s to 570, mainly due to the identification work by me and J. Tuiskunen (Bergman and Jansson 1983, Tuiskunen and Lindeberg 1986). -

Volume 2, Chapter 12-19: Terrestrial Insects: Holometabola-Diptera
Glime, J. M. 2017. Terrestrial Insects: Holometabola – Diptera Nematocera 2. In: Glime, J. M. Bryophyte Ecology. Volume 2. 12-19-1 Interactions. Ebook sponsored by Michigan Technological University and the International Association of Bryologists. eBook last updated 19 July 2020 and available at <http://digitalcommons.mtu.edu/bryophyte-ecology2/>. CHAPTER 12-19 TERRESTRIAL INSECTS: HOLOMETABOLA – DIPTERA NEMATOCERA 2 TABLE OF CONTENTS Cecidomyiidae – Gall Midges ........................................................................................................................ 12-19-2 Mycetophilidae – Fungus Gnats ..................................................................................................................... 12-19-3 Sciaridae – Dark-winged Fungus Gnats ......................................................................................................... 12-19-4 Ceratopogonidae – Biting Midges .................................................................................................................. 12-19-6 Chironomidae – Midges ................................................................................................................................. 12-19-9 Belgica .................................................................................................................................................. 12-19-14 Culicidae – Mosquitoes ................................................................................................................................ 12-19-15 Simuliidae – Blackflies -

2 3 Influence of Invasive Palms on Terrestrial Arthropod Assemblages 4
1 1 2011. Biological Conservation 144: 518-525 2 http://www.sciencedirect.com/science/article/pii/S0006320710004404 3 4 Influence of invasive palms on terrestrial arthropod assemblages 5 in desert spring habitat 6 7 Jeffrey G. Holmquista,1,*, Jutta Schmidt-Gengenbacha,1, Michèle R. Slatonb,2 8 9 aUniversity of California San Diego, White Mountain Research Station, 3000 East Line 10 Street, Bishop, California, USA 93514 11 bDeath Valley National Park, Death Valley, California, USA 92328 12 13 *Corresponding author [email protected] 14 15 1current: University of California Los Angeles, Institute of the Environment and 16 Sustainability, White Mountain Research Center, 3000 East Line Street, Bishop, California, 17 USA 93514 18 2current: Inyo National Forest, 351 Pacu Lane, Suite 200, Bishop, California, USA 93514 19 20 21 22 23 2 1 ABSTRACT 2 Invasive plants can eliminate native flora and ultimately have negative indirect effects on 3 fauna and the functional ecology of ecosystems, but understanding of these cascading effects 4 on arthropod assemblages is poor. Desert spring habitats are small, isolated landscape 5 elements that are literal oases for flora and fauna and support high diversity assemblages; 6 invasive palms can colonize desert springs and form monocultures. In an effort to understand 7 effects of these invasive trees on higher terrestrial trophic levels at springs, we contrasted 8 assemblage structure of terrestrial arthropods in native vegetation versus invasive palm 9 habitat. We sampled arthropods in paired palm and native habitat at ten springs in Death 10 Valley National Park, California, USA, during both spring and fall growing seasons using 11 suction sampling. -
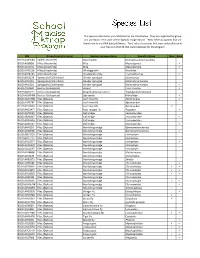
Species List
The species collected in your Malaise trap are listed below. They are organized by group and are listed in the order of the 'Species Image Library'. ‘New’ refers to species that are brand new to our DNA barcode library. 'Rare' refers to species that were only collected in your trap out of all 68 that were deployed for the program. BIN Group (scientific name) Species Common Name Scientific Name New Rare BOLD:AAA9584 Spiders (Araneae) Dwarf spider Dismodicus decemoculatus BOLD:ACI8838 Mites (Arachnida) Mite Mesostigmata BOLD:ACJ2455 Mites (Arachnida) Mite Mesostigmata BOLD:AAF9236 Mites (Arachnida) Whirligig mite Anystidae BOLD:ACR4249 Mites (Arachnida) Prostigmatic mite Trombidiformes BOLD:ACL6239 Springtails (Collembola) Slender springtail Entomobrya BOLD:ACI5504 Springtails (Collembola) Slender springtail Entomobryomorpha BOLD:AAI1229 Springtails (Collembola) Slender springtail Entomobryomorpha BOLD:AAI2228 Beetles (Coleoptera) Weevil Curculionidae BOLD:AAB3221 Beetles (Coleoptera) Striped ambrosia beetle Trypodendron lineatum BOLD:ACM3484 Beetles (Coleoptera) Sap beetle Nitidulidae BOLD:AAG4788 Flies (Diptera) Leaf miner fly Agromyzidae BOLD:AAP6787 Flies (Diptera) Leaf miner fly Agromyzidae BOLD:ACU8660 Flies (Diptera) Leaf miner fly Agromyzidae BOLD:AAG2477 Flies (Diptera) Root-maggot fly Pegomya BOLD:AAP9039 Flies (Diptera) Gall midge Cecidomyiidae BOLD:ABA6427 Flies (Diptera) Gall midge Cecidomyiidae BOLD:ACD6956 Flies (Diptera) Gall midge Cecidomyiidae BOLD:ABA6517 Flies (Diptera) Gall midge Cecidomyiidae BOLD:AAI4193 -

Dipterists Digest: Contents 1988–2021
Dipterists Digest: contents 1988–2021 Latest update at 12 August 2021. Includes contents for all volumes from Series 1 Volume 1 (1988) to Series 2 Volume 28(2) (2021). For more information go to the Dipterists Forum website where many volumes are available to download. Author/s Year Title Series Volume Family keyword/s EDITOR 2021 Corrections and changes to the Diptera Checklist (46) 2 28 (2): 252 LIAM CROWLEY 2021 Pandivirilia melaleuca (Loew) (Diptera, Therevidae) recorded from 2 28 (2): 250–251 Therevidae Wytham Woods, Oxfordshire ALASTAIR J. HOTCHKISS 2021 Phytomyza sedicola (Hering) (Diptera, Agromyzidae) new to Wales and 2 28 (2): 249–250 Agromyzidae a second British record Owen Lonsdale and Charles S. 2021 What makes a ‘good’ genus? Reconsideration of Chromatomyia Hardy 2 28 (2): 221–249 Agromyzidae Eiseman (Diptera, Agromyzidae) ROBERT J. WOLTON and BENJAMIN 2021 The impact of cattle on the Diptera and other insect fauna of a 2 28 (2): 201–220 FIELD temperate wet woodland BARRY P. WARRINGTON and ADAM 2021 The larval habits of Ophiomyia senecionina Hering (Diptera, 2 28 (2): 195–200 Agromyzidae PARKER Agromyzidae) on common ragwort (Jacobaea vulgaris) stems GRAHAM E. ROTHERAY 2021 The enigmatic head of the cyclorrhaphan larva (Diptera, Cyclorrhapha) 2 28 (2): 178–194 MALCOLM BLYTHE and RICHARD P. 2021 The biting midge Forcipomyia tenuis (Winnertz) (Diptera, 2 28 (2): 175–177 Ceratopogonidae LANE Ceratopogonidae) new to Britain IVAN PERRY 2021 Aphaniosoma melitense Ebejer (Diptera, Chyromyidae) in Essex and 2 28 (2): 173–174 Chyromyidae some recent records of A. socium Collin DAVE BRICE and RYAN MITCHELL 2021 Recent records of Minilimosina secundaria (Duda) (Diptera, 2 28 (2): 171–173 Sphaeroceridae Sphaeroceridae) from Berkshire IAIN MACGOWAN and IAN M. -

National Park Service
Communities in Freshwater Coastal Rock Pools of Lake Superior, with a Focus on Chironomidae (Diptera) A Dissertation SUBMITTED TO THE FACULTY OF UNIVERSITY OF MINNESOTA BY Alexander Taurus Egan IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY Advisor: Leonard C. Ferrington, Jr. May 2014 © Alexander Taurus Egan 2014 Acknowledgements Projects of this size are rarely accomplished without the assistance and support of many people. Primarily, my advisor, Len Ferrington, has been a great source of guidance and enthusiasm. My committee, Jacques Finlay, Ralph Holzenthal, and Roger Moon, have raised the bar considerably by pushing, pulling and steering me toward being a better scientist. Friends and colleagues in the Chironomidae Research Group have made my graduate experience a time I will remember fondly, with Alyssa Anderson, Will Bouchard and Jessica Miller sharing in the successes, misfortunes, and minor but important goals that come with the territory. In particular, Petra Kranzfelder often filled the roles of peer advisor and sounding board for ideas both brilliant and ridiculous. The National Park service has been very generous in many ways, and specific thanks go to Brenda Moraska Lafrançois and Jay Glase, who provided early development and direction for this project. My colleagues Mark Edlund from the Science Museum of Minnesota and Toben Lafrançois from the Science Museum and Northland College have consistently offered excellent ecological advice on what the data mean, often acting as de facto advisors. Without support from Isle Royale National Park this project would not have been possible. In particular, the technical advice, equipment loans, and logistical assistance from Paul Brown, Rick Damstra, Joan Elias, and Mark Romanski were invaluable. -

Assessment of the Efficacy of BTI Larvicide, Vectobac 12AS® (BTI AM65-52) Against “Nuisance” Flies at Water Treatment Plants in North West England
Assessment of the Efficacy of BTI Larvicide, VectoBac 12AS® (BTI AM65-52) Against “Nuisance” Flies at Water Treatment Plants in North West England. Standreck Wenjere School of Environment and Life Sciences University of Salford, Manchester, UK. Submitted in Partial Fulfilment of the Requirements of the Degree of Master of Science (by Research), 2017. 1 Table of Contents Page List of tables………………………………………………………………………………………. 5 List of figures……………………………………………………………………………………... 7 Acknowledgements……………………………………………………………………………….. 9 Abbreviations and key words……………………………………………….…………………….. 10 Abstract…………………………………………………………………………………………… 11 Chapter 1. Introduction 1.1 General overview…………………………………………………………………………. 13 1.2 Water industry: companies and regulators……………………………………………….. 14 1.3 Advent of wastewater treatment: environment and public health………………………… 15 1.4 General outline of wastewater treatment…………………………………………………. 21 1.5 Ecology of bacteria beds……………………………………………………………….…. 22 1.5.1 Bacteria…………………………………………………………………………………. 23 1.5.2 Algae, Fungi and protozoa……………………………………………….……………... 23 1.5.3 Annelid worms……………………………………………….…………………………. 24 1.5.4 Nematode worms….…………………………………….………………………………. 24 1.5.5 Molluscs – snails and slugs……………………………………………….…………….. 25 1.5.6 Nematoceran diptera……………………………………………………………………. 25 1.6 Common sewage-associated fly species….………………………………………………. 26 1.6.1 Sylvicola fenestralis…………………………………………………………………….. 27 1.6.2 Metriocnemus eurynotus………………………………………………….…………….. 29 1.6.3 Limnophyes minimus………………………………………………….………………… -

De Eurasian Lakes with Respect to Temperature and Continentality: Development and Application of New Chironomid-Based Climate-Inference Models in Northern Russia
Quaternary Science Reviews 30 (2011) 1122e1141 Contents lists available at ScienceDirect Quaternary Science Reviews journal homepage: www.elsevier.com/locate/quascirev The distribution and abundance of chironomids in high-latitude Eurasian lakes with respect to temperature and continentality: development and application of new chironomid-based climate-inference models in northern Russia A.E. Self a,b,*, S.J. Brooks a, H.J.B. Birks b,c,d, L. Nazarova e, D. Porinchu f, A. Odland g, H. Yang b, V.J. Jones b a Department of Entomology, Natural History Museum, Cromwell Road, London SW7 5BD, UK b Environmental Change Research Centre, Department of Geography, University College London, Gower Street, London WC1E 6BT, UK c Department of Biology, Bjerknes Centre for Climate Research, University of Bergen, P.O. Box 7803, N-5020 Bergen, Norway d School of Geography and the Environment, University of Oxford, South Parks Road, Oxford OX1 3QY, UK e Alfred Wegener Institute for Polar and Marine Research, Telegrafenberg A 43, 14473 Potsdam, Germany f Department of Geography, The Ohio State University, 1036 Derby Hall, 154 N. Oval Mall, Columbus, OH 43210, USA g Institute of Environmental Studies, Telemark University College, N-3800 Bø, Norway article info abstract Article history: The large landmass of northern Russia has the potential to influence global climate through amplification Received 10 June 2010 of climate change. Reconstructing climate in this region over millennial timescales is crucial for Received in revised form understanding the processes that affect the global climate system. Chironomids, preserved in lake 18 January 2011 sediments, have the potential to produce high resolution, low error, quantitative summer air temperature Accepted 19 January 2011 reconstructions. -

Fly Times Issue 64
FLY TIMES ISSUE 64, Spring, 2020 Stephen D. Gaimari, editor Plant Pest Diagnostics Branch California Department of Food & Agriculture 3294 Meadowview Road Sacramento, California 95832, USA Tel: (916) 738-6671 FAX: (916) 262-1190 Email: [email protected] Welcome to the latest issue of Fly Times! This issue is brought to you during the Covid-19 pandemic, with many of you likely cooped up at home, with insect collections worldwide closed for business! Perhaps for this reason this issue is pretty heavy, not just with articles but with images. There were many submissions to the Flies are Amazing! section and the Dipterists Lairs! I hope you enjoy them! Just to touch on an error I made in the Fall issue’s introduction… In outlining the change to “Spring” and “Fall” issues, instead of April and October issues, I said “But rest assured, I WILL NOT produce Fall issues after 20 December! Nor Spring issues after 20 March!” But of course I meant no Spring issues after 20 June! Instead of hitting the end of spring, I used the beginning. Oh well… Thank you to everyone for sending in such interesting articles! I encourage all of you to consider contributing articles that may be of interest to the Diptera community, or for larger manuscripts, the Fly Times Supplement series. Fly Times offers a great forum to report on research activities, to make specimen requests, to report interesting observations about flies or new and improved methods, to advertise opportunities for dipterists, to report on or announce meetings relevant to the community, etc., with all the digital images you wish to provide. -

Diptera: Chironomidae) in the Sediment of Plešné Lake (The Bohemian Forest, Czech Republic): Palaeoenvironmental Implications
Biologia, Bratislava, 61/Suppl. 20: S401—S411, 2006 Section Zoology DOI: 10.2478/s11756-007-0076-6 Holocene subfossil chironomid stratigraphy (Diptera: Chironomidae) in the sediment of Plešné Lake (the Bohemian Forest, Czech Republic): Palaeoenvironmental implications Jolana Tátosová 1,JosefVeselý2 & Evžen Stuchlík3 1Institute for Environmental Studies, Faculty of Science, Charles University in Prague, Benátská 2,CZ-12801 Prague, Czech Republic; e-mail: [email protected] 2Czech Geological Survey, Geologická 6,CZ-15200 Prague 5, Czech Republic 3Hydrobiological Station, Institute for Environmental Studies, Charles University in Prague, P.O. Box 47,CZ-38801 Blatná, Czech Republic Abstract: A faunal record of chironomid remains was analyzed in the upper 280 cm of a 543 cm long sediment core from Plešné jezero (Plešné Lake), the Bohemian Forest (Šumava, B¨ohmerwald), Czech Republic. The chronology of the sediment was established by means of 5 AMS-dated plant macroremains. The resolution of individual 3-cm sediment layers is ∼115 years and the analyzed upper 280 cm of the sediment core represent 10.4 cal. ka BP. As the results of DCA show, two marked changes were recorded in the otherwise relatively stable Holocene chironomid composition: (1) at the beginning of the Holocene (ca. 10.4–10.1 cal. ka BP) only oligotrophic and cold-adapted taxa (Diamesa sp.,M.insignilobus-type, H. grimshawi-type) were present in the chironomid assemblages, clearly reflecting a cool climate oscillation during the Preboreal period, and (2) during an event dated in the interval 1540–1771 AD, when most taxa vanished entirely and only Zavrelimyia sp. and Procladius sp.