The Concise Guide to PHARMACOLOGY 2015/16: Catalytic Receptors
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Katalog 2015 Cover Paul Lin *Hinweis Förderung.Indd
Product List 2015 WE LIVE SERVICE Certificates quartett owns two productions sites that are certified according to EN ISO 9001:2008 Quality management systems - Requirements EN ISO 13485:2012 + AC:2012 Medical devices - Quality management systems - Requirements for regulatory purposes GMP Conformity Our quality management guarantees products of highest quality! 2 Foreword to the quartett product list 2015 quartett Immunodiagnostika, Biotechnologie + Kosmetik Vertriebs GmbH welcomes you as one of our new business partners as well as all of our previous loyal clients. You are now member of quartett´s worldwide customers. First of all we would like to introduce ourselves to you. Founded as a family-run company in 1986, quartett ensures for more than a quarter of a century consistent quality of products. Service and support of our valued customers are our daily businesses. And we will continue! In the end 80´s quartett offered radioimmunoassay and enzyme immunoassay kits from different manufacturers in the USA. In the beginning 90´s the company changed its strategy from offering products for routine diagnostic to the increasing field of research and development. Setting up a production plant in 1997 and a second one in 2011 supported this decision. The company specialized its product profile in the field of manufacturing synthetic peptides for antibody production, peptides such as protease inhibitors, biochemical reagents and products for histology, cytology and immunohistology. All products are exclusively manufactured in Germany without outsourcing any production step. Nowadays, we expand into all other diagnostic and research fields and supply our customers in universities, government institutes, pharmaceutical and biotechnological companies, hospitals, and private doctor offices. -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

TRAIL and Cardiovascular Disease—A Risk Factor Or Risk Marker: a Systematic Review
Journal of Clinical Medicine Review TRAIL and Cardiovascular Disease—A Risk Factor or Risk Marker: A Systematic Review Katarzyna Kakareko 1,* , Alicja Rydzewska-Rosołowska 1 , Edyta Zbroch 2 and Tomasz Hryszko 1 1 2nd Department of Nephrology and Hypertension with Dialysis Unit, Medical University of Białystok, 15-276 Białystok, Poland; [email protected] (A.R.-R.); [email protected] (T.H.) 2 Department of Internal Medicine and Hypertension, Medical University of Białystok, 15-276 Białystok, Poland; [email protected] * Correspondence: [email protected] Abstract: Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a pro-apoptotic protein showing broad biological functions. Data from animal studies indicate that TRAIL may possibly contribute to the pathophysiology of cardiomyopathy, atherosclerosis, ischemic stroke and abdomi- nal aortic aneurysm. It has been also suggested that TRAIL might be useful in cardiovascular risk stratification. This systematic review aimed to evaluate whether TRAIL is a risk factor or risk marker in cardiovascular diseases (CVDs) focusing on major adverse cardiovascular events. Two databases (PubMed and Cochrane Library) were searched until December 2020 without a year limit in accor- dance to the PRISMA guidelines. A total of 63 eligible original studies were identified and included in our systematic review. Studies suggest an important role of TRAIL in disorders such as heart failure, myocardial infarction, atrial fibrillation, ischemic stroke, peripheral artery disease, and pul- monary and gestational hypertension. Most evidence associates reduced TRAIL levels and increased TRAIL-R2 concentration with all-cause mortality in patients with CVDs. It is, however, unclear Citation: Kakareko, K.; whether low TRAIL levels should be considered as a risk factor rather than a risk marker of CVDs. -

Mtorc1-Independent Autophagy Regulates Receptor Tyrosine Kinase Phosphorylation in Colorectal Cancer Cells Via an Mtorc2-Mediated Mechanism
Cell Death and Differentiation (2017) 24, 1045–1062 & 2017 Macmillan Publishers Limited, part of Springer Nature. All rights reserved 1350-9047/17 www.nature.com/cdd mTORC1-independent autophagy regulates receptor tyrosine kinase phosphorylation in colorectal cancer cells via an mTORC2-mediated mechanism Aikaterini Lampada1,2, James O’Prey3, Gyorgy Szabadkai4, Kevin M Ryan3, Daniel Hochhauser*,2,5 and Paolo Salomoni*,1,5 The intracellular autophagic degradative pathway can have a tumour suppressive or tumour-promoting role depending on the stage of tumour development. Upon starvation or targeting of oncogenic receptor tyrosine kinases (RTKs), autophagy is activated owing to the inhibition of PI3K/AKT/mTORC1 signalling pathway and promotes survival, suggesting that autophagy is a relevant therapeutic target in these settings. However, the role of autophagy in cancer cells where the PI3K/AKT/mTORC1 pathway is constitutively active remains partially understood. Here we report a role for mTORC1-independent basal autophagy in regulation of RTK activation and cell migration in colorectal cancer (CRC) cells. PI3K and RAS-mutant CRC cells display basal autophagy levels despite constitutive mTORC1 signalling, but fail to increase autophagic flux upon RTK inhibition. Inhibition of basal autophagy via knockdown of ATG7 or ATG5 leads to decreased phosphorylation of several RTKs, in particular c-MET. Internalised c-MET colocalised with LAMP1-negative, LC3-positive vesicles. Finally, autophagy regulates c-MET phosphorylation via an mTORC2- dependent -

Angiopoietin 4 (ANGPT4) (NM 015985) Human Untagged Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for SC304373 Angiopoietin 4 (ANGPT4) (NM_015985) Human Untagged Clone Product data: Product Type: Expression Plasmids Product Name: Angiopoietin 4 (ANGPT4) (NM_015985) Human Untagged Clone Tag: Tag Free Symbol: ANGPT4 Synonyms: ANG3; ANG4 Vector: pCMV6-XL5 E. coli Selection: Ampicillin (100 ug/mL) Cell Selection: None This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 3 Angiopoietin 4 (ANGPT4) (NM_015985) Human Untagged Clone – SC304373 Fully Sequenced ORF: >OriGene sequence for NM_015985 edited CAGGCAAGCCTGGCCACTGTTGGCTGCAGCAGGACATCCCAGGCACAGCCCCTAGGGCTC TGAGCAGACATCCCTCGCCATTGACACATCTTCAGATGCTCTCCCAGCTAGCCATGCTGC AGGGCAGCCTCCTCCTTGTGGTTGCCACCATGTCTGTGGCTCAACAGACAAGGCAGGAGG CGGATAGGGGCTGCGAGACACTTGTAGTCCAGCACGGCCACTGTAGCTACACCTTCTTGC TGCCCAAGTCTGAGCCCTGCCCTCCGGGGCCTGAGGTCTCCAGGGACTCCAACACCCTCC AGAGAGAATCACTGGCCAACCCACTGCACCTGGGGAAGTTGCCCACCCAGCAGGTGAAAC AGCTGGAGCAGGCACTGCAGAACAACACGCAGTGGCTGAAGAAGCTAGAGAGGGCCATCA AGACGATCTTGAGGTCGAAGCTGGAGCAGGTCCAGCAGCAAATGGCCCAGAATCAGACGG CCCCCATGCTAGAGCTGGGCACCAGCCTCCTGAACCAGACCACTGCCCAGATCCGCAAGC TGACCGACATGGAGGCTCAGCTCCTGAACCAGACATCAAGAATGGATGCCCAGATGCCAG AGACCTTTCTGTCCACCAACAAGCTGGAGAACCAGCTGCTGCTACAGAGGCAGAAGCTCC AGCAGCTTCAGGGCCAAAACAGCGCGCTCGAGAAGCGGTTGCAGGCCCTGGAGACCAAGC -
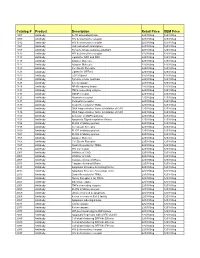
Comprehensive Product List
Catalog # Product Description Retail Price OEM Price 1007 Antibody IL-1R associated kinase 225/100ug 125/100ug 1009 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1012 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1021 Antibody JAK activated transcription 225/100ug 125/100ug 1107 Antibody Tyrosine kinase substrate p62DOK 225/100ug 125/100ug 1112 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1113 Antibody Ligand for DR4 and DR5 225/100ug 125/100ug 1115 Antibody Adapter Molecule 225/100ug 125/100ug 1117 Antibody Adapter Molecule 225/100ug 125/100ug 1120 Antibody Cell Death Receptor 225/100ug 125/100ug 1121 Antibody Ligand for GFRa-2 225/100ug 125/100ug 1123 Antibody CCR3 ligand 225/100ug 125/100ug 1125 Antibody Tyrosine kinase substrate 225/100ug 125/100ug 1128 Antibody A new caspase 225/100ug 125/100ug 1129 Antibody NF-kB inducing kinase 225/100ug 125/100ug 1131 Antibody TNFa converting enzyme 225/100ug 125/100ug 1133 Antibody GDNF receptor 225/100ug 125/100ug 1135 Antibody Neurturin receptor 225/100ug 125/100ug 1137 Antibody Persephin receptor 225/100ug 125/100ug 1139 Antibody Death Receptor for TRAIL 225/100ug 125/100ug 1141 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1148 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1150 Antibody Activator of MAPK pathway 225/100ug 125/100ug 1151 Antibody Apoptosis Signal-regulation Kinase 225/100ug 125/100ug 1156 Antibody FLICE inhibitory protein 225/100ug 125/100ug 1158 Antibody Cell Death Receptor 225/100ug 125/100ug 1159 -

The Role of Overexpressed Apolipoprotein AV in Insulin-Resistant Hepatocytes
Hindawi BioMed Research International Volume 2020, Article ID 3268505, 13 pages https://doi.org/10.1155/2020/3268505 Research Article The Role of Overexpressed Apolipoprotein AV in Insulin-Resistant Hepatocytes Wang Zhao,1 Yaqiong Liu,1 Xiaobo Liao,2 and Shuiping Zhao 1 1Department of Cardiovascular Medicine, e Second Xiangya Hospital, Central South University, No. 139 Middle Renmin Road, Changsha, Hunan 410011, China 2Department of Cardiovascular Surgery, e Second Xiangya Hospital, Central South University, No. 139 Middle Renmin Road, Changsha, Hunan 410011, China Correspondence should be addressed to Shuiping Zhao; [email protected] Received 31 January 2019; Revised 16 July 2019; Accepted 16 August 2019; Published 21 April 2020 Academic Editor: Kui Li Copyright © 2020 Wang Zhao et al. *is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In this paper, we sought to explore the relationship between apolipoprotein AV (APOAV) overexpression and insulin resistance in hepatocytes. *e insulin-resistant HepG2 cell model was constructed, and then, APOAV-overexpressed HepG2 cells (B-M) were induced by infecting with a recombinant adenovirus vector. Microarray data were developed from B-M samples compared with negative controls (A-con), and the microarray data were analyzed by bioinformatic methods. APOAV-overexpression induced 313 upregulated genes and 563 downregulated ones in B-M sample. *e differentially expressed genes (DEGs) were significantly classified in fat digestion and absorption pathway. Protein-protein interaction network was constructed, and AGTR1 (angiotensin II receptor type 1) and P2RY2 (purinergic receptor P2Y, G-protein coupled 2) were found to be the significant nodes closely related with G-protein related signaling. -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

Dcr2 (Decoy Receptor 2, TRAIL-R4, TRUNDD)/Fc Chimera Human, Recombinant Expressed in Mouse NSO Cells
DcR2 (Decoy Receptor 2, TRAIL-R4, TRUNDD)/Fc Chimera Human, Recombinant Expressed in mouse NSO cells Product Number D9813 Product Description Reagents Recombinant human DcR2 (TRAIL-R4, TRUNDD) is a DcR2 is supplied as approximately 100 mg of protein chimeric protein expressed in mouse NSO cells. The lyophilized from a 0.2 mm filtered solution in phosphate 1, 2 extracellular domain of human DcR2 is fused to the buffered saline. carboxy-terminal 6X histidine-tagged Fc portion of human IgG1 by a polypeptide linker. Mature Preparation Instructions recombinant human DcR2 is a disulfide-linked Reconstitute the contents of the vial using sterile homodimeric protein. The reduced DcR2 monomer has phosphate-buffered saline (PBS) containing at least a molecular mass of approximately 44.2 kDa. Due to 0.1% human serum albumin or bovine serum albumin. glycosylation, recombinant human DcR2 migrates as an Prepare a stock solution of no less than 50 mg/ml. approximately 70-80 kDa protein in SDS-PAGE under reducing conditions. Storage/Stability Store at -20°C. Upon reconstitution, store at 2°-8°C for Apoptosis or programmed cell death is induced in cells one month. For extended storage, freeze in working by a group of death domain-containing receptors aliquots. Repeated freezing and thawing is not including TNFR1, Fas, DR3, DR4, and DR5. Binding of recommended. ligand to these receptors sends signals that activate members of the caspase family of proteases. The Product Profile signals ultimately cause the degradation of chromo- DcR2 is measured by its ability to neutralize apoptosis somal DNA by activating DNase. of mouse L929 cells treated with 50 ng/ml TRAIL. -

Protein Tyrosine Kinases: Their Roles and Their Targeting in Leukemia
cancers Review Protein Tyrosine Kinases: Their Roles and Their Targeting in Leukemia Kalpana K. Bhanumathy 1,*, Amrutha Balagopal 1, Frederick S. Vizeacoumar 2 , Franco J. Vizeacoumar 1,3, Andrew Freywald 2 and Vincenzo Giambra 4,* 1 Division of Oncology, College of Medicine, University of Saskatchewan, Saskatoon, SK S7N 5E5, Canada; [email protected] (A.B.); [email protected] (F.J.V.) 2 Department of Pathology and Laboratory Medicine, College of Medicine, University of Saskatchewan, Saskatoon, SK S7N 5E5, Canada; [email protected] (F.S.V.); [email protected] (A.F.) 3 Cancer Research Department, Saskatchewan Cancer Agency, 107 Wiggins Road, Saskatoon, SK S7N 5E5, Canada 4 Institute for Stem Cell Biology, Regenerative Medicine and Innovative Therapies (ISBReMIT), Fondazione IRCCS Casa Sollievo della Sofferenza, 71013 San Giovanni Rotondo, FG, Italy * Correspondence: [email protected] (K.K.B.); [email protected] (V.G.); Tel.: +1-(306)-716-7456 (K.K.B.); +39-0882-416574 (V.G.) Simple Summary: Protein phosphorylation is a key regulatory mechanism that controls a wide variety of cellular responses. This process is catalysed by the members of the protein kinase su- perfamily that are classified into two main families based on their ability to phosphorylate either tyrosine or serine and threonine residues in their substrates. Massive research efforts have been invested in dissecting the functions of tyrosine kinases, revealing their importance in the initiation and progression of human malignancies. Based on these investigations, numerous tyrosine kinase inhibitors have been included in clinical protocols and proved to be effective in targeted therapies for various haematological malignancies. -

Biolegend.Com
Mechanisms of Cell Death TRAIL (TNFSF10) TNF-α Death Receptor 4 (TNFRSF10A/TRAIL-R1) Death Receptor 5 Zombie Dyes (TNFRSF10B/TRAIL-R2) Propidium Iodide (PI) BAT1, TIM-4 TNF RI (TNFRSF1A) 7-Amino-Actinomycin (7-AAD) MER TNF RII (TNFRSF1B) FAS-L GAS6 (TNFSF6/CD178) TRAIL (TNFSF10) Apoptotic Cell Death Domain Zombie Dyes Phosphatidylserine K63 Ubiquitin NH2 Removal ICAM3? ROCK1 NH CD14 2 Eat-Me Signals FAS Death Inducing Cytoskeletal Rearrangement, (TNFRSF6/CD95) Signaling Complex (DISC) TRADD Cytoskeletal Rearrangement, TRADD Decoy Receptor 2 FADD (TNFRSF10D/TRAIL-R4) Actomysin Contraction Engulfment RIP1 TWEAK RIP1 oxLDL (TNFSF12) FADD CIAP1/2 K63 Ubiquitination Blebbing CD36 Death Receptor 3 TWEAK (TNFSF12) PI FADD (TNFRSF25, APO-3) 7-AAD TRAF1 FADD Procaspase 8,10 TRAF 3 Phagocyte FLIP PANX1 Macrophage Monocyte Neutrophil Dendritic Cell Fibroblast Mast Cell Procaspase 8,10 NF-kB TWEAK-R (TNFRSF12A/Fn14) Find-Me Signals Lysophosphocholine C Caspase 8,10 TRAF5 TRAF2 Sphingosine-1-Phosphate G2A? Nucleotides A Decoy TRAIL Receptor R1 (TNFRSF23) Bid Cell Survival ATP, UTP Decoy TRAIL Receptor R2 (TNFRSF22) Sphingosine-1 TRADD Phosphate Receptor Decoy Receptor 1 (TNFRSF10C/TRAIL-R3) Procaspase 3 Proliferation RIP1 G P2y2 t-Bid Bcl-2 T Chemotaxis, Caspase 3 Bcl-2-xL, MCL-1 ? ICAD RIP1 Engulfment Degradation Bax, Bak Oligomerization TRADD Death Receptor 6 Extracellular ATP Bacterial pore-forming toxins TRAIL (TNFSF10) ICAD (TNFRSF21) Monosodium urate crystals Cholesterol crystals Death Receptor DNA Fragmentation Cholera toxin B, Mitochondria -

WO 2018/027204 Al 08 February 2018 (08.02.2018) W !P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/027204 Al 08 February 2018 (08.02.2018) W !P O PCT (51) International Patent Classification: only): F. HOFFMANN-LA ROCHE AG [CH/CH]; Gren- C07K 16/28 (2006.01) A61K 39/00 (2006.01) zacherstrasse 124, 4070 Basel (CH). (21) International Application Number: (72) Inventor; and PCT/US20 17/045642 (71) Applicant: HARRIS, Seth [US/US]; c/o Genentech, Inc., 1 DNA Way, South San Francisco, California 94080 (US). (22) International Filing Date: 04 August 2017 (04.08.2017) (72) Inventors: LAZAR, Greg; c/o Genentech, Inc., 1 DNA Way, South San Francisco, California 94080 (US). YANG, (25) Filing Language: English Yanli; c/o Genentech, Inc., 1 DNA Way, South San Fran (26) Publication Language: English cisco, California 94080 (US). CHRISTENSEN, Erin H.; c/ o Genentech, Inc., 1 DNA Way, South San Francisco, Cali (30) Priority Data: fornia 94080 (US). HANG, Julie; 6606 Wisteria Way, San 62/371,671 05 August 2016 (05.08.2016) US Jose, California 95 129 (US). KIM, Jeong; c/o Genentech, (71) Applicant (for all designated States except AL, AT, BA, BE, Inc., 1 DNA Way, South San Francisco, California 94080 BG, CH, CN, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, (US). HR, HU, IE, IN, IS, IT, LT, LU, LV, MC, MK, MT, NL, (74) Agent: JONES, Kevin et al; Morrison & Foerster LLP, NO, PL, PT RO, RS, SE, SI, SK, SM, TR): GENENTECH, 425 Market Street, San Francisco, California 94105-2482 INC.