Bile Acids and Their Derivatives As Vitamin D Receptor Agonists:Molecular Mechanism and Biological Actions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Suitability of Immobilized Systems for Microbiological Degradation of Endocrine Disrupting Compounds
molecules Review Suitability of Immobilized Systems for Microbiological Degradation of Endocrine Disrupting Compounds Danuta Wojcieszy ´nska , Ariel Marchlewicz and Urszula Guzik * Institute of Biology, Biotechnology and Environmental Protection, Faculty of Natural Science, University of Silesia in Katowice, Jagiello´nska28, 40-032 Katowice, Poland; [email protected] (D.W.); [email protected] (A.M.) * Correspondence: [email protected]; Tel.: +48-3220-095-67 Academic Editors: Urszula Guzik and Danuta Wojcieszy´nska Received: 10 September 2020; Accepted: 25 September 2020; Published: 29 September 2020 Abstract: The rising pollution of the environment with endocrine disrupting compounds has increased interest in searching for new, effective bioremediation methods. Particular attention is paid to the search for microorganisms with high degradation potential and the possibility of their use in the degradation of endocrine disrupting compounds. Increasingly, immobilized microorganisms or enzymes are used in biodegradation systems. This review presents the main sources of endocrine disrupting compounds and identifies the risks associated with their presence in the environment. The main pathways of degradation of these compounds by microorganisms are also presented. The last part is devoted to an overview of the immobilization methods used for the purposes of enabling the use of biocatalysts in environmental bioremediation. Keywords: EDCs; hormones; degradation; immobilization; microorganisms 1. Introduction The development of modern tools for the separation and identification of chemical substances has drawn attention to the so-called micropollution of the environment. Many of these contaminants belong to the emergent pollutants class. According to the Stockholm Convention they are characterized by high persistence, are transported over long distances in the environment through water, accumulate in the tissue of living organisms and can adversely affect them [1]. -

A Left/Right Comparison of Twice-Daily Calcipotriol Ointment and Calcitriol Ointment in Patients with Psoriasis: the Effect on Keratinocyte Subpopulations
Acta Derm Venereol 2004; 84: 195–200 INVESTIGATIVE REPORT A Left/Right Comparison of Twice-Daily Calcipotriol Ointment and Calcitriol Ointment in Patients with Psoriasis: The Effect on Keratinocyte Subpopulations Mannon E.J. FRANSSEN, Gys J. DE JONGH, Piet E.J. VAN ERP and Peter C.M. VAN DE KERKHOF Department of Dermatology, University Medical Centre Nijmegen, The Netherlands Vitamin D3 analogues are a first-line treatment of Calcipotriol (Daivonex1,50mg/g ointment, Leo chronic plaque psoriasis, but so far, comparative clinical Pharmaceutical Products, Denmark) has been investi- studies on calcipotriol and calcitriol ointment are sparse, gated intensively during the last decade, and has proven and in particular no comparative studies are available on to be a valuable tool in the management of chronic cell biological effects of these compounds in vivo. Using plaque psoriasis. A review by Ashcroft et al. (1), based on flow cytometric assessment, we investigated whether these a large number of randomized controlled trials, showed compounds had different effects on the composition and that calcipotriol was at least as effective as potent DNA synthesis of epidermal cell populations responsible topical corticosteroids, 1a,-25-dihydroxycholecalciferol for the psoriatic phenotype. For 8 weeks, 20 patients with (calcitriol), short-contact dithranol, tacalcitol and coal psoriasis vulgaris were treated twice daily with calcipo- tar. Recently, Scott et al. (2) presented an overview of triol and calcitriol ointment in a left/right comparative studies on the use of calcipotriol ointment in the study. Before and after treatment, clinical assessment of management of psoriasis. They reconfirmed the super- target lesions was performed, together with flow cyto- ior efficacy of a twice-daily calcipotriol ointment metric analysis of epidermal subpopulations with respect regimen to the treatments as mentioned above, and to keratin (K) 10, K6, vimentin and DNA distribution. -

Abstract Mahapatra, Debabrata
ABSTRACT MAHAPATRA, DEBABRATA. Vitamin D Receptor and Xenobiotic Interactions: Insights into the diversity and complexity of molecular interactions and their outcomes (Under the direction of Seth W. Kullman). The etiology of a significant number of human diseases including hypertension, cardiovascular disease, diabetes, obesity and cancer are in part associated with exposures to environmental contaminants. Recent trends in toxicological research indicate a growing interest in chemicals capable of disrupting the endocrine system. These chemicals comprise a range of natural and synthetic molecules that interact with nuclear hormone receptors altering signaling pathways regulating adverse effects in multiple organ systems, tissue and cell types. While the interaction of endocrine disrupting xenobiotics with nuclear receptors such as the Estrogen receptor (ER), Thyroid receptor (TR), Glucocorticoid receptor (GR) and Androgen receptor (AR) has been studied in detail, similar research involving xenobiotic interactions with the Vitamin D receptor (VDR) has remained underexplored. This dissertation examines the role of vitamin d receptor as a potential target of xenobiotic induced endocrine disruption and provides new insights into the possible mechanisms underlying the complex molecular interactions between VDR and select VDR agonists and antagonists. Chapter 1 tests the usefulness and importance of orthogonal assays for validation and confirmation of activity profiles of chemicals generated by the qHTS (Quantitative Highthroughput Testing) -

Part I Biopharmaceuticals
1 Part I Biopharmaceuticals Translational Medicine: Molecular Pharmacology and Drug Discovery First Edition. Edited by Robert A. Meyers. © 2018 Wiley-VCH Verlag GmbH & Co. KGaA. Published 2018 by Wiley-VCH Verlag GmbH & Co. KGaA. 3 1 Analogs and Antagonists of Male Sex Hormones Robert W. Brueggemeier The Ohio State University, Division of Medicinal Chemistry and Pharmacognosy, College of Pharmacy, Columbus, Ohio 43210, USA 1Introduction6 2 Historical 6 3 Endogenous Male Sex Hormones 7 3.1 Occurrence and Physiological Roles 7 3.2 Biosynthesis 8 3.3 Absorption and Distribution 12 3.4 Metabolism 13 3.4.1 Reductive Metabolism 14 3.4.2 Oxidative Metabolism 17 3.5 Mechanism of Action 19 4 Synthetic Androgens 24 4.1 Current Drugs on the Market 24 4.2 Therapeutic Uses and Bioassays 25 4.3 Structure–Activity Relationships for Steroidal Androgens 26 4.3.1 Early Modifications 26 4.3.2 Methylated Derivatives 26 4.3.3 Ester Derivatives 27 4.3.4 Halo Derivatives 27 4.3.5 Other Androgen Derivatives 28 4.3.6 Summary of Structure–Activity Relationships of Steroidal Androgens 28 4.4 Nonsteroidal Androgens, Selective Androgen Receptor Modulators (SARMs) 30 4.5 Absorption, Distribution, and Metabolism 31 4.6 Toxicities 32 Translational Medicine: Molecular Pharmacology and Drug Discovery First Edition. Edited by Robert A. Meyers. © 2018 Wiley-VCH Verlag GmbH & Co. KGaA. Published 2018 by Wiley-VCH Verlag GmbH & Co. KGaA. 4 Analogs and Antagonists of Male Sex Hormones 5 Anabolic Agents 32 5.1 Current Drugs on the Market 32 5.2 Therapeutic Uses and Bioassays -

Evolution of the Bile Salt Nuclear Receptor FXR in Vertebrates
Supplemental Material can be found at: http://www.jlr.org/cgi/content/full/M800138-JLR200/DC1 Evolution of the bile salt nuclear receptor FXR in vertebrates † † † †† †† Erica J. Reschly,* Ni Ai, Sean Ekins, ,§,** William J. Welsh, Lee R. Hagey, Alan F. Hofmann, and Matthew D. Krasowski1,* † Department of Pathology,* University of Pittsburgh, Pittsburgh, PA 15261; Department of Pharmacology, University of Medicine and Dentistry of New Jersey, Robert Wood Johnson Medical School, Piscataway, NJ 08854; Collaborations in Chemistry,§ Jenkintown, PA 19046; Department of Pharmaceutical Sciences,** †† University of Maryland, Baltimore, MD 21202; and Department of Medicine, University of California-San Diego, San Diego, CA 92093-0063 Abstract Bile salts, the major end metabolites of cho- Bile salts are water-soluble, amphipathic end metabolites lesterol, vary significantly in structure across vertebrate of cholesterol that facilitate intestinal absorption of lipids species, suggesting that nuclear receptors binding these (1), enhance proteolytic cleavage of dietary proteins (2), Downloaded from molecules may show adaptive evolutionary changes. We com- and have potent antimicrobial activity in the small intestine pared across species the bile salt specificity of the major (3). In addition, bile salt signaling via nuclear hormone re- transcriptional regulator of bile salt synthesis, the farnesoid X receptor (FXR). We found that FXRs have changed speci- ceptors (NHRs) is important for bile salt homeostasis (4). ficity for primary bile salts across species by altering the Bile salts have not been detected in invertebrate animals. shape and size of the ligand binding pocket. In particular, In contrast to steroid hormones and vitamins, whose struc- the ligand binding pockets of sea lamprey (Petromyzon marinus) tures tend to be strongly conserved, bile salts exhibit www.jlr.org and zebrafish (Danio rerio) FXRs, as predicted by homology marked structural diversity across species (5–7). -

Eldecalcitol Is More Effective for Promoting Osteogenesis Than Alfacalcidol in Cyp27b1-Knockout Mice
bioRxiv preprint doi: https://doi.org/10.1101/349837; this version posted June 18, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Eldecalcitol is more effective for promoting osteogenesis than alfacalcidol in Cyp27b1-knockout Mice Short title: Osteogenic effect of eldecalcitol Yoshihisa Hirota1,2*¶, Kimie Nakagawa2¶, Keigo Isomoto2¶, Toshiyuki Sakaki3, Noboru Kubodera4, Maya Kamao2, Naomi Osakabe5, Yoshitomo Suhara6, Toshio Okano2* 1 Laboratory of Biochemistry, Department of Bioscience and Engineering, College of Systems Engineering and Science, Shibaura Institute of Technology, 307 Fukasaku, Minuma-ku, Saitama 337-8570, Japan 2 Laboratory of Hygienic Sciences, Kobe Pharmaceutical University, 4-19-1 Motoyamakita-machi, Higashinada-ku, Kobe 658-8558, Japan 3 Department of Pharmaceutical Engineering, Faculty of Engineering, Toyama Prefectural University, Kurokawa, Imizu, Toyama 939-0398, Japan 4 International Institute of Active Vitamin D Analogs, 35-6, Sankeidai, Mishima, Shizuoka 411-0017, Japan 5 Food and Nutrition Laboratory, Department of Bioscience and Engineering, College of Systems Engineering and Science, Shibaura Institute of Technology, 307 Fukasaku, Minuma-ku, Saitama 337-8570, Japan 6 Laboratory of Organic Synthesis and Medicinal Chemistry, Department of Bioscience and Engineering, College of Systems Engineering and Science, Shibaura Institute of Technology, 307 Fukasaku, Minuma-ku, Saitama 337-8570, Japan ¶ These authors contributed equally to this work. * Corresponding authors: Yoshihisa Hirota Tel.: +81-48-7201-6037; Fax: +81-48-7201-6011; E-mail: hirotay@ shibaura-it.ac.jp Toshio Okano Tel.: (81) 78-441-7524; Fax: (81) 78-441-7524; E-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/349837; this version posted June 18, 2018. -

A New Role for Vitamin D Receptor Activation in Chronic Kidney Disease
A NEW ROLE FOR VITAMIN D RECEPTOR ACTIVATION IN CHRONIC KIDNEY DISEASE José M Valdivielso 1, Jorge Cannata-Andía2, Blai Coll 1 and Elvira Fernández 1 1Laboratorio de Nefrología Experimental, Hospital Universitario Arnau de Vilanova, IRBLLEIDA, REDinREN ISCIII Lleida, Spain. 2Servicio de Metabolismo Óseo y Mineral. Hospital Universitario Central de Asturias. Instituto “Reina Sofía” de Investigación. REDinREN ISCIII. Universidad de Oviedo Running Title: New role of VDRA Correspondence to: Dr. José M Valdivielso, Laboratorio de Nefrología Experimental, IRBLLEIDA Hospital Universitari Arnau de Vilanova Rovira Roure 80 25198 Lleida, Spain. Phone: +34 973003650 Fax: +34 973702213 e-mail: [email protected] Abstract Vitamin D has proven to be much more than a simple ‘calcium hormone’. The fact that the vitamin D receptor has been found in cells not related to mineral metabolism supports that statement. The interest of nephrologists in Vitamin D and its effects beyond mineral metabolism has increased over the last few years, evidencing the importance of this so-called ‘sunshine hormone’. In the present review, we highlight the most recent developments in the traditional use of vitamin D in chronic kidney disease (CKD) patients, namely the control of secondary hyperparathyroidism (sHPT). Furthermore, we also explore the data available regarding the new possible therapeutic uses of vitamin D for the treatment of other complications present in CKD patients, such as vascular calcification, left ventricular hypertrophy or proteinuria. Finally, some still scarce but very promising data regarding a possible role of vitamin D in kidney transplant patients are also reviewed. The available data point to a potential beneficial effect of vitamin D in CKD patients beyond the control of mineral metabolism. -
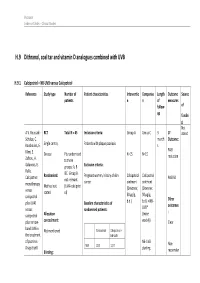
H.9 Dithranol, Coal Tar and Vitamin D Analogues Combined with UVB
Psoriasis Evidence Tables – Clinical Studies H.9 Dithranol, coal tar and vitamin D analogues combined with UVB H.9.1 Calcipotriol + NB-UVB versus Calcipotriol Reference Study type Number of Patient characteristics Interventio Compariso Length Outcome Source patients n n of measures follow- of up fundin g Not A.V. Roussaki- RCT Total N = 45 Inclusion criteria: Group A Group C 3 1º stated Schulze, C. month Outcome: Kouskoukis, E. Single centre, Patients with plaque psoriasis s Klimi, E. PASI Greece Pts randomised N=15 N=15 reduction Zafirou, A. to three Galanous, E. groups: A, B Exclusion criteria: Rallis. &C. Group B Calcipotriol Randomised: Pregnant women, history of skin Calcipotriol Calcipotriol PASI 50 not relevant cancer ointment ointment monotherapy Method not (UVA+calcipotri versus (Dovonex; (Dovonex stated ol) 50 µg/g, 50 µg/g, calcipotriol Other plus UVA1 Baseline characteristics of b.d.) b.d.) + NB- UVB* outcomes versus randomised patients: : calcipotriol Allocation (twice plus narrow- concealment: weekly) Clear band UVB in Calcipotriol Calcipotriol + Not mentioned the treatment NB-UVB of psoriasis. NB-UVB Non- Drugs Exptl. M/F 12/3 12/3 starting Blinding: responder Psoriasis Evidence Tables – Clinical Studies Cl in. Res , dose 80% 31(5/6):169- Not mentioned Age 44.93±6.48 49.53±22.01 MED and 174.2005 inc. by 20% Skin type 0/11/3/1 2/5/6/2 I/II/III/IV every 3 REFID: Washout sessions ROUSSAKISCH period: ULZE2005 90 days if using systemic *Cosmetico therapy, 30 days , 10 lamps if using topicals Helarium B1, 100 W each. -

1,25-Dihydroxyvitamin D3-Induced Genes in Osteoblasts
1,25-DIHYDROXYVITAMIN D3-INDUCED GENES IN OSTEOBLASTS: UNCOVERING NEW FUNCTIONS FOR MENINGIOMA 1 AND SEMAPHORIN 3B IN SKELETAL PHYSIOLOGY by XIAOXUE ZHANG Submitted in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Thesis advisor: Paul N. MacDonald Department of Pharmacology CASE WESTERN RESERVE UNIVERSITY May 2009 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of _____________________________________________________ candidate for the ______________________degree *. (signed)_______________________________________________ (chair of the committee) ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. I dedicate this thesis to my mother and father for their lifelong love, encouragement and sacrifice TABLE OF CONTENTS Table of Contents ii List of Tables iii List of Figures iv Acknowledgements vii Abbreviations x Abstract xiii Chapter I Introduction 1 Chapter II Meningioma 1 (MN1) is a 1,25-dihydroxyvitamin D3- 44 induced transcription coactivator that promotes osteoblast proliferation, motility, differentiation, and function Chapter III Semaphorin 3B (SEMA3B) is a 1,25- 108 dihydroxyvitamin D3-induced gene in osteoblasts that promotes -

Is Calcifediol Better Than Cholecalciferol for Vitamin D Supplementation?
Osteoporosis International (2018) 29:1697–1711 https://doi.org/10.1007/s00198-018-4520-y REVIEW Is calcifediol better than cholecalciferol for vitamin D supplementation? J. M. Quesada-Gomez1,2 & R. Bouillon3 Received: 22 February 2018 /Accepted: 28 March 2018 /Published online: 30 April 2018 # International Osteoporosis Foundation and National Osteoporosis Foundation 2018 Abstract Modest and even severe vitamin D deficiency is widely prevalent around the world. There is consensus that a good vitamin D status is necessary for bone and general health. Similarly, a better vitamin D status is essential for optimal efficacy of antiresorptive treatments. Supplementation of food with vitamin D or using vitamin D supplements is the most widely used strategy to improve the vitamin status. Cholecalciferol (vitamin D3) and ergocalciferol (vitamin D2)arethemostwidelyused compounds and the relative use of both products depends on historical or practical reasons. Oral intake of calcifediol (25OHD3) rather than vitamin D itself should also be considered for oral supplementation. We reviewed all publications dealing with a comparison of oral cholecalciferol with oral calcifediol as to define the relative efficacy of both compounds for improving the vitamin D status. First, oral calcifediol results in a more rapid increase in serum 25OHD compared to oral cholecalciferol. Second, oral calcifediol is more potent than cholecalciferol, so that lower dosages are needed. Based on the results of nine RCTs comparing physiologic doses of oral cholecalciferol with oral calcifediol, calcifediol was 3.2-fold more potent than oral chole- calciferol. Indeed, when using dosages ≤ 25 μg/day, serum 25OHD increased by 1.5 ± 0.9 nmol/l for each 1 μgcholecalciferol, whereas this was 4.8 ± 1.2 nmol/l for oral calcifediol. -

(12) Patent Application Publication (10) Pub. No.: US 2011/001412.6 A1 Evans Et Al
US 2011 0014126A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2011/001412.6 A1 Evans et al. (43) Pub. Date: Jan. 20, 2011 (54) USE OF VITAMIND RECEPTORAGONISTS (60) Provisional application No. 60/985,972, filed on Nov. AND PRECURSORS TO TREAT FIBROSS 6, 2007. (76) Inventors: Ronald M. Evans, La Jolla, CA Publication Classification (US); Michael Downes, San Diego, CA (US); Christopher Liddle, (51) Int. Cl. New South Wales (AU): A 6LX 3/59 (2006.01) Nanthakumar Subramaniam, A6IPL/I6 (2006.01) New South Wales (AU); Caroline CI2O 1/02 (2006.01) Flora Samer, Geneva (CH) A61R 49/00 (2006.01) CI2N 5/071 (2010.01) Correspondence Address: (52) U.S. Cl. ............. 424/9.2: 514/167; 435/29: 435/375 KLARQUIST SPARKMAN, LLP 121 S.W. SALMONSTREET, SUITE 1600 (57) ABSTRACT PORTLAND, OR 97204 (US) This application relates to methods of treating, preventing, (21) Appl. No.: 12/772,981 and ameliorating fibrosis, such as fibrosis of the liver. In particular, the application relates to methods of using a vita (22) Filed: May 3, 2010 min D receptor agonist (Such as vitamin D. Vitamin Dana logs, vitamin D precursors, and vitamin D receptor agonists Related U.S. Application Data precursors) for the treatment of liver fibrosis. Also disclosed (63) Continuation-in-part of application No. 12/266,513, are methods for screening for agents that treat, prevent, and filed on Nov. 6, 2008. ameliorate fibrosis. Stellate Cells Liver RR1,3 AR, ERa ERR1,2,3, AR, ERa CNF GR, MR CNF RARa, GR, MR NF4g Rab NF4ag RARa,b, NOR1 a, RH1 TRa,b WDR NURR1 NOR1 RORa,b,g RORag CAR Receptor SF-1 FXRa,b FXRa,b epissertoup. -

Paricalcitol Versus Calcitriol in the Treatment of Secondary Hyperparathyroidism
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Kidney International, Vol. 63 (2003), pp. 1483–1490 Paricalcitol versus calcitriol in the treatment of secondary hyperparathyroidism STUART M. SPRAGUE,FRANCISCO LLACH,MICHAEL AMDAHL,CAROL TACCETTA, and DANIEL BATLLE Division of Nephrology/Hypertension and Department of Medicine, Northwestern University Feinberg School of Medicine, Evanston and Chicago, Illinois; Georgetown University Medical School, Washington, D.C.; and Abbott Laboratories, North Chicago, Illinois Paricalcitol versus calcitriol in the treatment of secondary hyper- Osteitis fibrosa cystica, resulting from poorly con- parathyroidism. trolled secondary hyperparathyroidism, remains a com- Background. Management of secondary hyperparathyroid- mon manifestation of renal osteodystrophy and causes ism has included the use of active vitamin D or vitamin D ana- logs for the suppression of parathyroid hormone (PTH) secre- significant morbidity in patients with chronic renal fail- tion. Although, these agents are effective, therapy is frequently ure [1, 2]. Decreased renal production of calcitriol (1,25 limited by hypercalcemia, hyperphosphatemia, and/or eleva- vitamin D ), hypocalcemia, and hyperphosphatemia are ϫ 3 tions in the calcium-phosphorus (Ca P) product. In clinical the major contributing factors to the development of studies, paricalcitol was shown to be effective at reducing PTH concentrations without causing significant hypercalcemia or secondary hyperparathyroidism [1, 3–5]. Calcitriol is syn- hyperphosphatemia as compared to placebo. A comparative thesized from previtamin D3 by hydroxylation on carbons study was undertaken in order to determine whether paricalci- 25 and 1 in the liver and kidney, respectively [6]. The tol provides a therapeutic advantage to calcitriol.