A Metabolomics Analysis to Define the Source of Heme
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Molecular Expression, Characterization and Mechanism of ALAS2 Gain-Of-Function Mutants Vassili Tchaikovskii, Robert J
Tchaikovskii et al. Molecular Medicine (2019) 25:4 Molecular Medicine https://doi.org/10.1186/s10020-019-0070-9 RESEARCHARTICLE Open Access Molecular expression, characterization and mechanism of ALAS2 gain-of-function mutants Vassili Tchaikovskii, Robert J. Desnick and David F. Bishop* Abstract Background: X-linked protoporphyria (XLP) (MIM 300752) is an erythropoietic porphyria due to gain-of-function mutations in the last exon (Ducamp et al., Hum Mol Genet 22:1280-88, 2013) of the erythroid-specific aminolevulinate synthase gene (ALAS2). Five ALAS2 exon 11 variants identified by the NHBLI Exome sequencing project (p.R559H, p.E565D, p.R572C, p.S573F and p.Y586F) were expressed, purified and characterized in order to assess their possible contribution to XLP. To further characterize the XLP gain-of-function region, five novel ALAS2 truncation mutations (p.P561X, p.V562X, p.H563X, p.E569X and p.F575X) were also expressed and studied. Methods: Site-directed mutagenesis was used to generate ALAS2 mutant clones and all were prokaryotically expressed, purified to near homogeneity and characterized by protein and enzyme kinetic assays. Standard deviations were calculated for 3 or more assay replicates. Results: The five ALAS2 single nucleotide variants had from 1.3- to 1.9-fold increases in succinyl-CoA Vmax and 2- to 3-fold increases in thermostability suggesting that most could be gain-of-function modifiers of porphyria instead of causes. One SNP (p.R559H) had markedly low purification yield indicating enzyme instability as the likely cause for XLSA in an elderly patient with x-linked sideroblastic anemia. The five novel ALAS2 truncation mutations had increased Vmax values for both succinyl-CoA and glycine substrates (1.4 to 5.6-fold over wild-type), while the Kms for both substrates were only modestly changed. -

Pseudouridine Synthase 1: a Site-Specific Synthase Without Strict Sequence Recognition Requirements Bryan S
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central Published online 18 November 2011 Nucleic Acids Research, 2012, Vol. 40, No. 5 2107–2118 doi:10.1093/nar/gkr1017 Pseudouridine synthase 1: a site-specific synthase without strict sequence recognition requirements Bryan S. Sibert and Jeffrey R. Patton* Department of Pathology, Microbiology and Immunology, University of South Carolina, School of Medicine, Columbia, SC 29208 USA Received May 20, 2011; Revised October 19, 2011; Accepted October 22, 2011 ABSTRACT rRNA and snRNA and requires Dyskerin or its homologs Pseudouridine synthase 1 (Pus1p) is an unusual (Cbf5p in yeast for example) and RNP cofactors [most site-specific modification enzyme in that it can often H/ACA small nucleolar ribonucleoprotein particles modify a number of positions in tRNAs and can rec- (snoRNPs)] that enable one enzyme to recognize many ognize several other types of RNA. No consensus different sites for modification on different substrates recognition sequence or structure has been identi- (17–25). The other pathway for É formation employs fied for Pus1p. Human Pus1p was used to determine site-specific É synthases that require no cofactors to rec- which structural or sequence elements of human ognize and modify the RNA substrate. A number of en- tRNASer are necessary for pseudouridine ()) forma- zymes have been identified in this pathway and are grouped in six families that all share a common basic tion at position 28 in the anticodon stem-loop (ASL). Ser structure (4). It is safe to say the cofactor ‘guided’ pathway Some point mutations in the ASL stem of tRNA has received a great deal of attention because of its simi- had significant effects on the levels of modification larity to aspects of RNA editing, but the site-specific and compensatory mutation, to reform the base pseudouridine synthases accomplish the same task, on pair, restored a wild-type level of ) formation. -

GATA Transcription Factors Directly Regulate the Parkinson's
GATA transcription factors directly regulate the Parkinson’s disease-linked gene ␣-synuclein Clemens R. Scherzer*†‡, Jeffrey A. Grass§, Zhixiang Liao*, Imelda Pepivani*, Bin Zheng*, Aron C. Eklund*, Paul A. Ney¶, Juliana Ngʈ, Meghan McGoldrick*, Brit Mollenhauer*, Emery H. Bresnick†§**, and Michael G. Schlossmacher*†ʈ†† *Center for Neurologic Diseases, Harvard Medical School and Brigham and Women’s Hospital, Cambridge, MA 02139; §Department of Pharmacology, University of Wisconsin School of Medicine and Public Health, Madison, WI 53706; ¶Department of Biochemistry, St. Jude Children’s Research Hospital, Memphis, TN 38105; and ʈDivision of Neurosciences, Ottawa Health Research Institute, University of Ottawa, Ottawa, ON, Canada K1H 8M5 Edited by Gregory A. Petsko, Brandeis University, Waltham, MA, and approved May 27, 2008 (received for review March 13, 2008) Increased ␣-synuclein gene (SNCA) dosage due to locus multipli- Results cation causes autosomal dominant Parkinson’s disease (PD). Vari- SNCA mRNA and Protein Are Highly Abundant in Human and Mouse ation in SNCA expression may be critical in common, genetically Erythroid Cells. Relative SNCA mRNA abundance was determined complex PD but the underlying regulatory mechanism is unknown. by comparing the SNCA mRNA abundance in each target tissue to We show that SNCA and the heme metabolism genes ALAS2, FECH, the calibrator Universal Human Reference RNA (Fig. 1A). SNCA and BLVRB form a block of tightly correlated gene expression in 113 mRNA abundance was high in whole blood from human donors samples of human blood, where SNCA naturally abounds (vali- (relative abundance 10 {range 6.6–15.3}) with very high levels in ؊ ؊ ؊ -؋ 10 11, 1.8 ؋ 10 10, and 6.6 ؋ 10 5). -

TITLE PAGE Oxidative Stress and Response to Thymidylate Synthase
Downloaded from molpharm.aspetjournals.org at ASPET Journals on October 2, 2021 -Targeted -Targeted 1 , University of of , University SC K.W.B., South Columbia, (U.O., Carolina, This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
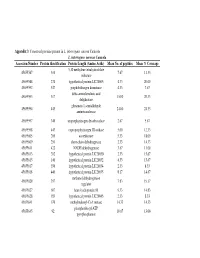
Appendix 3 and 4
Appendix 3 : Conserved proteins present in L. interrogans serovar Canicola L. interrogans serovar Canicola Accession Number Protein identification Protein Length (Amino Acids) Mean No. of peptides Mean % Coverage 5,10 methylene tetrahydrofolate 45655587 310 7.67 11.33 reductase 45655588 274 hypothetical protein LIC20005 4.33 20.00 45655592 547 porphobilinogen deaminase 4.33 7.67 delta-aminolevulinic acid 45655593 317 15.00 20.33 dehydratase glutamate-1-semialdehyde 45655594 443 24.00 24.33 aminotransferase 45655597 340 uroporphyrinogen decarboxylase 2.67 5.67 45655598 443 coproporphyrinogen III oxidase 5.00 12.33 45655605 200 azoreductase 5.33 18.00 45655609 251 short-chain dehydrogenase 2.33 14.33 45655611 422 NADH dehydrogenase 3.67 11.00 45655613 202 hypothetical protein LIC20030 2.33 15.67 45655615 140 hypothetical protein LIC20032 4.33 15.67 45655617 356 hypothetical protein LIC20034 2.33 8.33 45655618 440 hypothetical protein LIC20035 8.17 14.67 methanol dehydrogenase 45655620 357 7.83 19.17 regulator 45655627 607 heat shock protein 90 9.33 14.83 45655628 189 hypothetical protein LIC20045 2.33 8.33 45655641 670 methylmalonyl-CoA mutase 14.33 14.33 phosphoribosyl-ATP 45655645 92 10.67 13.00 pyrophosphatase 3-oxoacyl-(acyl-carrier protein) 45655647 254 4.00 17.00 reductase 45655648 77 acyl carrier protein 3.67 9.00 45655661 171 hypothetical protein LIC20078 5.00 26.00 4-hydroxybenzoyl-CoA 45655663 142 3.33 11.67 thioesterase S-adenosyl-L-homocysteine 45655666 436 13.00 19.67 hydrolase B12-dependent methionine 45655668 1247 42.67 21.67 -

Case Reports
CASE REPORTS The 5’ untranslated region (UTR) of ALAS2 mRNA A mutation in the iron-responsive element of contains a cis-regulatory iron-responsive element (IRE) ALAS2 is a modifier of disease severity in a that confers iron-dependent posttranscriptional regula - patient suffering from CLPX associated erythro - tion by the iron regulatory proteins (IRP). 7 IRE muta - poietic protoporphyria tions are known to cause human diseases. IRE muta - tions in ferritin L mRNA cause hereditary hyperferritine - Porphyrias are a group of eight genetically distinct dis - mia with cataract syndrome (HHCS) (OMIM orders, each resulting from a partial deficiency or gain-of- #600886), 8-9 and a single point mutation in the IRE of function of a specific enzyme in the heme biosynthetic ferritin H is responsible for an autosomal dominant iron 1 pathway. Porphyrias are inherited as autosomal domi - overload phenotype (OMIM #615517). 10 These cases 2 nant, autosomal recessive or X-linked traits. suggest a possible role for IRE mutations in the ALAS2 Erythropoietic protoporphyria (EPP) is a constitutive gene in modifying the severity of hematologic diseases. hematological disorder characterized by protoporphyrin Here, we describe an 18 year-old Caucasian female IX (PPIX) accumulation in erythrocytes and other tissues proband (III:2, Figure 1A) referred to the French Center resulting in acute skin photosensitivity, mild microcytic of Porphyria because of early onset (9 months) acute anemia, and rarely, severe liver disease. The majority of photosensitivity characterized by painfully phototoxic the patients with EPP present the autosomal EPP form reactions suggesting EPP. An incomplete genetic charac - (OMIM #177000) due to a partial deficiency of fer - terization of this case was previously reported to harbor rochelatase (FECH), the last enzyme of the heme biosyn - a gain of function in the mitochondrial unfoldase gene, thetic pathway. -

Figure S1. HAEC ROS Production and ML090 NOX5-Inhibition
Figure S1. HAEC ROS production and ML090 NOX5-inhibition. (a) Extracellular H2O2 production in HAEC treated with ML090 at different concentrations and 24 h after being infected with GFP and NOX5-β adenoviruses (MOI 100). **p< 0.01, and ****p< 0.0001 vs control NOX5-β-infected cells (ML090, 0 nM). Results expressed as mean ± SEM. Fold increase vs GFP-infected cells with 0 nM of ML090. n= 6. (b) NOX5-β overexpression and DHE oxidation in HAEC. Representative images from three experiments are shown. Intracellular superoxide anion production of HAEC 24 h after infection with GFP and NOX5-β adenoviruses at different MOIs treated or not with ML090 (10 nM). MOI: Multiplicity of infection. Figure S2. Ontology analysis of HAEC infected with NOX5-β. Ontology analysis shows that the response to unfolded protein is the most relevant. Figure S3. UPR mRNA expression in heart of infarcted transgenic mice. n= 12-13. Results expressed as mean ± SEM. Table S1: Altered gene expression due to NOX5-β expression at 12 h (bold, highlighted in yellow). N12hvsG12h N18hvsG18h N24hvsG24h GeneName GeneDescription TranscriptID logFC p-value logFC p-value logFC p-value family with sequence similarity NM_052966 1.45 1.20E-17 2.44 3.27E-19 2.96 6.24E-21 FAM129A 129. member A DnaJ (Hsp40) homolog. NM_001130182 2.19 9.83E-20 2.94 2.90E-19 3.01 1.68E-19 DNAJA4 subfamily A. member 4 phorbol-12-myristate-13-acetate- NM_021127 0.93 1.84E-12 2.41 1.32E-17 2.69 1.43E-18 PMAIP1 induced protein 1 E2F7 E2F transcription factor 7 NM_203394 0.71 8.35E-11 2.20 2.21E-17 2.48 1.84E-18 DnaJ (Hsp40) homolog. -

REPORT C-Terminal Deletions in the ALAS2 Gene Lead to Gain of Function and Cause X-Linked Dominant Protoporphyria Without Anemia Or Iron Overload
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector REPORT C-Terminal Deletions in the ALAS2 Gene Lead to Gain of Function and Cause X-linked Dominant Protoporphyria without Anemia or Iron Overload Sharon D. Whatley,1,9 Sarah Ducamp,2,3,9 Laurent Gouya,2,3 Bernard Grandchamp,3,4 Carole Beaumont,3 Michael N. Badminton,1 George H. Elder,1 S. Alexander Holme,5 Alexander V. Anstey,5 Michelle Parker,6 Anne V. Corrigall,6 Peter N. Meissner,6 Richard J. Hift,6 Joanne T. Marsden,7 Yun Ma,8 Giorgina Mieli-Vergani,8 Jean-Charles Deybach,2,3,* and Herve´ Puy2,3 All reported mutations in ALAS2, which encodes the rate-regulating enzyme of erythroid heme biosynthesis, cause X-linked sideroblastic anemia. We describe eight families with ALAS2 deletions, either c.1706-1709 delAGTG (p.E569GfsX24) or c.1699-1700 delAT (p.M567EfsX2), resulting in frameshifts that lead to replacement or deletion of the 19–20 C-terminal residues of the enzyme. Prokaryotic expression studies show that both mutations markedly increase ALAS2 activity. These gain-of-function mutations cause a previously unrecognized form of porphyria, X-linked dominant protoporphyria, characterized biochemically by a high proportion of zinc-proto- porphyrin in erythrocytes, in which a mismatch between protoporphyrin production and the heme requirement of differentiating erythroid cells leads to overproduction of protoporphyrin in amounts sufficient to cause photosensitivity and liver disease. Each of the seven inherited porphyrias results from a partial mutations in about 7% of EPP families, of which about deficiency of an enzyme of heme biosynthesis. -
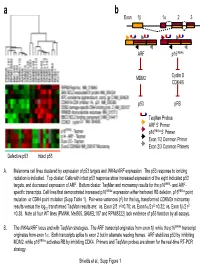
Taqman Probes ARF 5' Primer P16ink4a 5' Primer Exon 1/2
a b Exon 1β 1α 23 ARF p16INK4a MDM2 Cyclin D CDK4/6 p53 pRB TaqMan Probes ARF 5’ Primer p16INK4a 5’ Primer Exon 1/2 Common Primer Exon 2/3 Common Primers Defective p53 Intact p53 A. Melanoma cell lines clustered by expression of p53 targets and INK4a/ARF expression. The p53 response to ionizing radiation is indicated. Top cluster: Cells with intact p53 response show increased expression of the eight indicated p53 targets, and decreased expression of ARF. Bottom cluster: TaqMan and microarray results for the p16INK4- and ARF- specific transcripts. Cell lines that demonstrated increased p16INK4a expression either harbored RB deletion, p16INK4a point 2 mutation or CDK4 point mutation (Supp Table 1). Pair-wise variances (r ) for the log2 transformed CDKN2a microarray 2 2 2 results versus the log10 transformed TaqMan results are: vs. Exon 2/3 r =0.78; vs. Exon1α/2 r =0.32; vs. Exon 1β/2 r =0.38. Note all four WT lines (PMWK, Mel505, SKMEL187 and RPMI8322) lack evidence of p53 function by all assays. B. The INK4a/ARF locus and with TaqMan strategies. The ARF transcript originates from exon 1β while the p16INK4a transcript originates from exon 1α. Both transcripts splice to exon 2 but in alternate reading frames. ARF stabilizes p53 by inhibiting MDM2, while p16INK4a activates RB by inhibiting CDK4. Primers and TaqMan probes are shown for the real-time RT-PCR strategy. Shields et al., Supp Figure 1 SKMEL 28 U01 24h SKMEL WM2664 U01 48h WM2664 U01 24h 24 U01 48h SKMEL 24 U01 24h SKMEL 24 Untreated SKMEL 24 DMSO 48h SKMEL 24 DMSO 24h SKMEL WM2664 DMSO 24h WM2664 DMSO 48h WM2644 Untreated 28 Untreated SKMEL 28 DMSO 48h SKMEL 28 DMSO 24h SKMEL -3.00 -2.00 -1.00 0.00 1.00 2.00 3.00 relative to median expression Genes decreased by UO1 (863) HIF1A Hypoxia-inducible factor 1, alpha subunit NM_001530 RBBP8 Retinoblastoma binding protein 8 NM_002894 Homo sapiens, clone IMAGE:4337652, mRNA BC018676 EIF4EBP1 Eukaryotic translation initiation factor 4E binding protein EXOSC8 Exosome component 8 NM_181503 ENST00000321524 MCM7 MCM7 minichromosome maintenance deficient 7 (S. -

Q 297 Suppl USE
The following supplement accompanies the article Atlantic salmon raised with diets low in long-chain polyunsaturated n-3 fatty acids in freshwater have a Mycoplasma dominated gut microbiota at sea Yang Jin, Inga Leena Angell, Simen Rød Sandve, Lars Gustav Snipen, Yngvar Olsen, Knut Rudi* *Corresponding author: [email protected] Aquaculture Environment Interactions 11: 31–39 (2019) Table S1. Composition of high- and low LC-PUFA diets. Stage Fresh water Sea water Feed type High LC-PUFA Low LC-PUFA Fish oil Initial fish weight (g) 0.2 0.4 1 5 15 30 50 0.2 0.4 1 5 15 30 50 80 200 Feed size (mm) 0.6 0.9 1.3 1.7 2.2 2.8 3.5 0.6 0.9 1.3 1.7 2.2 2.8 3.5 3.5 4.9 North Atlantic fishmeal (%) 41 40 40 40 40 30 30 41 40 40 40 40 30 30 35 25 Plant meals (%) 46 45 45 42 40 49 48 46 45 45 42 40 49 48 39 46 Additives (%) 3.3 3.2 3.2 3.5 3.3 3.4 3.9 3.3 3.2 3.2 3.5 3.3 3.4 3.9 2.6 3.3 North Atlantic fish oil (%) 9.9 12 12 15 16 17 18 0 0 0 0 0 1.2 1.2 23 26 Linseed oil (%) 0 0 0 0 0 0 0 6.8 8.1 8.1 9.7 11 10 11 0 0 Palm oil (%) 0 0 0 0 0 0 0 3.2 3.8 3.8 5.4 5.9 5.8 5.9 0 0 Protein (%) 56 55 55 51 49 47 47 56 55 55 51 49 47 47 44 41 Fat (%) 16 18 18 21 22 22 22 16 18 18 21 22 22 22 28 31 EPA+DHA (% diet) 2.2 2.4 2.4 2.9 3.1 3.1 3.1 0.7 0.7 0.7 0.7 0.7 0.7 0.7 4 4.2 Table S2.