Investigation of Parkin Molecular Pathways
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
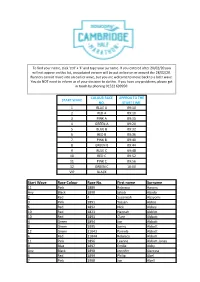
Start Wave Race Colour Race No. First Name Surname
To find your name, click 'ctrl' + 'F' and type your surname. If you entered after 20/02/20 you will not appear on this list, an updated version will be put online on or around the 28/02/20. Runners cannot move into an earlier wave, but you are welcome to move back to a later wave. You do NOT need to inform us of your decision to do this. If you have any problems, please get in touch by phoning 01522 699950. COLOUR RACE APPROX TO THE START WAVE NO. START TIME 1 BLUE A 09:10 2 RED A 09:10 3 PINK A 09:15 4 GREEN A 09:20 5 BLUE B 09:32 6 RED B 09:36 7 PINK B 09:40 8 GREEN B 09:44 9 BLUE C 09:48 10 RED C 09:52 11 PINK C 09:56 12 GREEN C 10:00 VIP BLACK Start Wave Race Colour Race No. First name Surname 11 Pink 1889 Rebecca Aarons Any Black 1890 Jakob Abada 2 Red 4 Susannah Abayomi 3 Pink 1891 Yassen Abbas 6 Red 1892 Nick Abbey 10 Red 1823 Hannah Abblitt 10 Red 1893 Clare Abbott 4 Green 1894 Jon Abbott 8 Green 1895 Jonny Abbott 12 Green 11043 Pamela Abbott 6 Red 11044 Rebecca Abbott 11 Pink 1896 Leanne Abbott-Jones 9 Blue 1897 Emilie Abby Any Black 1898 Jennifer Abecina 6 Red 1899 Philip Abel 7 Pink 1900 Jon Abell 10 Red 600 Kirsty Aberdein 6 Red 11045 Andrew Abery Any Black 1901 Erwann ABIVEN 11 Pink 1902 marie joan ablat 8 Green 1903 Teresa Ablewhite 9 Blue 1904 Ahid Abood 6 Red 1905 Alvin Abraham 9 Blue 1906 Deborah Abraham 6 Red 1907 Sophie Abraham 1 Blue 11046 Mitchell Abrams 4 Green 1908 David Abreu 11 Pink 11047 Kathleen Abuda 10 Red 11048 Annalisa Accascina 4 Green 1909 Luis Acedo 10 Red 11049 Vikas Acharya 11 Pink 11050 Catriona Ackermann -

View PDF Version
RSC Advances View Article Online PAPER View Journal | View Issue The trans/cis photoisomerization in hydrogen bonded complexes with stability controlled by Cite this: RSC Adv.,2018,8, 23698 substituent effects: 3-(6-aminopyridin-3-yl) acrylate case study† a a b a Adam Kwiatkowski, Beata Je˛drzejewska, Marek Jozefowicz,´ Izabela Grela c and Borys Osmia´ łowski * The association of aminopyridine-based acrylic acid and its salt was studied by NMR titration experiments. The AA (acceptor, acceptor) hydrogen-bonding pattern present in the salt forms a complex readily with a DD (donor, donor) hydrogen-bonding pattern of the substituted ureas even in polar and competitive environment. The double carbon–carbon bond in the acrylic acid derivative is subjected to photoisomerization. This is dependent on the association with substituted urea derivatives. The substituent in ureas influences the trans/cis isomerization kinetics and position of the photostationary Creative Commons Attribution-NonCommercial 3.0 Unported Licence. Received 9th April 2018 state. Two mechanisms that influence the photoisomerization were proposed. To the best of our Accepted 19th June 2018 knowledge, the trans/cis photoisomerization influenced by the substituent in such a hydrogen-bonding DOI: 10.1039/c8ra03042a pattern has not observed previously. It was shown that interaction with urea derivatives causes lowering rsc.li/rsc-advances of the trans-to-cis photoreaction rates. Introduction also in crystals15). Taking the proton transfer process into account some general remarks should be made. First, it is worth The urea moiety is one of the most popular supramolecular mentioning that carboxylic acids are self-complementary This article is licensed under a synthons.1–3 It forms double hydrogen-bonding but this feature (Fig. -

Genetic Analysis of Indel Markers in Three Loci Associated with Parkinson’S Disease
RESEARCH ARTICLE Genetic analysis of indel markers in three loci associated with Parkinson's disease Zhixin Huo1, Xiaoguang Luo2, Xiaoni Zhan1, Qiaohong Chu1, Qin Xu1, Jun Yao1, Hao Pang1* 1 Department of Forensic Genetics and Biology, China Medical University, Shenyang North New Area, Shenyang, P.R., China, 2 Department of Neurology, 1st Affiliated Hospital of China Medical University, Shenyang, P.R., China * [email protected] Abstract a1111111111 a1111111111 The causal mutations and genetic polymorphisms associated with susceptibility to Parkin- a1111111111 son's disease (PD) have been extensively described. To explore the potential contribution a1111111111 a1111111111 of insertion (I)/deletion (D) polymorphisms (indels) to the risk of PD in a Chinese population, we performed genetic analyses of indel loci in ACE, DJ-1, and GIGYF2 genes. Genomic DNA was extracted from venous blood of 348 PD patients and 325 age- and sex-matched controls without neurodegenerative disease. Genotyping of the indel loci was performed by fragment length analysis after PCR and DNA sequencing. Our results showed a statistically OPEN ACCESS significant association for both allele X (alleles without 5) vs. 5 (odds ratio = 1.378, 95% con- Citation: Huo Z, Luo X, Zhan X, Chu Q, Xu Q, Yao fidence interval = 1.112±1.708, P = 0.003) and genotype 5/X+X/X vs. 5/5 (odds ratio = J, et al. (2017) Genetic analysis of indel markers in three loci associated with Parkinson's disease. 1.681, 95% confidence interval = 1.174±2.407, P = 0.004) in the GIGYF2 locus; however, PLoS ONE 12(9): e0184269. https://doi.org/ no significant differences were detected for the ACE and DJ-1 indels. -

Characterisation of the Genomic Landscape of CRLF2‐Rearranged Acute Lymphoblastic Leukemia
Characterisation of the Genomic Landscape of CRLF2- rearranged Acute Lymphoblastic Leukemia Lisa J Russell1*, Lisa Jones1, Amir Enshaei1, Stefano Tonin1, Sarra L Ryan1, Jeyanthy Eswaran1 , Sirintra Nakjang2, Elli Papaemmanuil3,4, Jose M C Tubio4, Adele K Fielding5, Ajay Vora6, Peter J Campbell4, Anthony V Moorman1, and Christine J Harrison1 1 Leukaemia Research Cytogenetics Group, Northern Institute for Cancer Research, Newcastle University, Newcastle-upon-Tyne, UK 2 Bioinformatics Support Unit, Newcastle University, Newcastle-upon-Tyne, UK 3 Memorial Sloan Kettering Cancer Center, USA 4 Cancer Genome Project, Wellcome Trust Sanger Institute, Hinxton, UK 5 Research Department of Haemaoloty, UCL Cancer Institute, London, UK 6 Department of Haematology, Sheffield Children’s Hospital, Sheffield, UK; AVM and CJH contributed equally to this study Running Title – Genomic landscape of CRLF2 rearranged leukemia Correspondence to: Dr Lisa J Russell, Wolfson Childhood Cancer Research Centre, Northern Institute for Cancer Research, Newcastle University, Level 6, Herschel Building, Brewery Lane, Newcastle upon Tyne, NE1 7RU, [email protected]. Acknowledgements Support by: The Kay Kendall Leukaemia Fund, Leuka, European Haematology Association and Bloodwise (formerly Leukaemia and Lymphoma Research) This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process which may lead to differences between this version and the Version of Record. Please cite this article as an ‘Accepted Article’, doi: 10.1002/gcc.22439 This article is protected by copyright. All rights reserved. Genes, Chromosomes & Cancer Page 2 of 147 Deregulated expression of the type I cytokine receptor, CRLF2, is observed in 5-15% of precursor B-cell acute lymphoblastic leukaemia (B-ALL). -

17 - 19 October 2019
THE 12TH INTERNATIONAL SYMPOSIUM ON HEALTH INFORMATICS AND BIOINFORMATICS T G T A A A T G A G A A G T T G G G T A A A A A A A A T T T A A A A A A G G A A A A A A A G G G G G G G G A A A G G G G G G T T G G G G G G G T T T T G G T T G G G T T T G G T A A T T G G T T T A A A A A A A A T T T A A A A A A T T A A A A A A A T A T T T T T T A A A T T T T G T A G A A T T T A A 17 - 19 OCTOBER 2019 HIBIT 2019 ABSTRACT BOOK THE 12TH INTERNATIONAL SYMPOSIUM ON HEALTH INFORMATICS AND BIOINFORMATICS TABLE OF CONTENTS 1 8 WELCOME MESSAGE KEYNOTE LECTURERS 2 19 ORGANIZING COMMITEE INVITED SPEAKERS 3 29 SCIENTIFIC COMMITEE SELECTED ABSTRACTS FOR ORAL PRESENTATIONS 6 61 PROGRAM POSTER PRESENTATIONS Welcome Message The International Symposium on Health Informatics and Bioinformatics, (HIBIT), now in its twelfth year HIBIT 2019, aims to bring together academics, researchers and practitioners who work in these popular and fulfilling areas and to create the much- needed synergy among medical, biological and information technology sectors. HIBIT is one of the few conferences emphasizing such synergy. -

An Integrative Multi-Platform Analysis for Discovering Biomarkers Of
BMC Cancer BioMed Central Research article Open Access An integrative multi-platform analysis for discovering biomarkers of osteosarcoma Guodong Li†1,2, Wenjuan Zhang†3, Huazong Zeng*4, Lei Chen5, Wenjing Wang6, Jilong Liu4, Zhiyu Zhang1 and Zhengdong Cai*1,2 Address: 1Department of Orthopaedics, Tenth People's Hospital, Tongji University, Shanghai 200072, PR China, 2Department of Orthopaedics, Changhai Hospital, Second Military Medical University, Shanghai 200433, PR China, 3School of Life Sciences, Fudan University, Shanghai 200433, PR China, 4Shanghai Sensichip Co Ltd, Shanghai 200433, PR China, 5International Co-operation Laboratory on Signal Transduction, Eastern Hepatobiliary Surgery Institute, Second Military Medical University, Shanghai 200438, PR China and 6Shanghai Municipal Center for Disease Control & Prevention, Shanghai 200336, PR China Email: Guodong Li - [email protected]; Wenjuan Zhang - [email protected]; Huazong Zeng* - [email protected]; Lei Chen - [email protected]; Wenjing Wang - [email protected]; Jilong Liu - [email protected]; Zhiyu Zhang - [email protected]; Zhengdong Cai* - [email protected] * Corresponding authors †Equal contributors Published: 16 May 2009 Received: 6 December 2008 Accepted: 16 May 2009 BMC Cancer 2009, 9:150 doi:10.1186/1471-2407-9-150 This article is available from: http://www.biomedcentral.com/1471-2407/9/150 © 2009 Li et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: SELDI-TOF-MS (Surface Enhanced Laser Desorption/Ionization-Time of Flight-Mass Spectrometry) has become an attractive approach for cancer biomarker discovery due to its ability to resolve low mass proteins and high-throughput capability. -

"The Genecards Suite: from Gene Data Mining to Disease Genome Sequence Analyses". In: Current Protocols in Bioinformat
The GeneCards Suite: From Gene Data UNIT 1.30 Mining to Disease Genome Sequence Analyses Gil Stelzer,1,5 Naomi Rosen,1,5 Inbar Plaschkes,1,2 Shahar Zimmerman,1 Michal Twik,1 Simon Fishilevich,1 Tsippi Iny Stein,1 Ron Nudel,1 Iris Lieder,2 Yaron Mazor,2 Sergey Kaplan,2 Dvir Dahary,2,4 David Warshawsky,3 Yaron Guan-Golan,3 Asher Kohn,3 Noa Rappaport,1 Marilyn Safran,1 and Doron Lancet1,6 1Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, Israel 2LifeMap Sciences Ltd., Tel Aviv, Israel 3LifeMap Sciences Inc., Marshfield, Massachusetts 4Toldot Genetics Ltd., Hod Hasharon, Israel 5These authors contributed equally to the paper 6Corresponding author GeneCards, the human gene compendium, enables researchers to effectively navigate and inter-relate the wide universe of human genes, diseases, variants, proteins, cells, and biological pathways. Our recently launched Version 4 has a revamped infrastructure facilitating faster data updates, better-targeted data queries, and friendlier user experience. It also provides a stronger foundation for the GeneCards suite of companion databases and analysis tools. Improved data unification includes gene-disease links via MalaCards and merged biological pathways via PathCards, as well as drug information and proteome expression. VarElect, another suite member, is a phenotype prioritizer for next-generation sequencing, leveraging the GeneCards and MalaCards knowledgebase. It au- tomatically infers direct and indirect scored associations between hundreds or even thousands of variant-containing genes and disease phenotype terms. Var- Elect’s capabilities, either independently or within TGex, our comprehensive variant analysis pipeline, help prepare for the challenge of clinical projects that involve thousands of exome/genome NGS analyses. -

Gene Expression Responses in a Cellular Model of Parkinson's Disease
Gene Expression Responses in a Cellular Model of Parkinson's Disease Louis Beverly Brill II Manassas, Virginia B.A., Johns Hopkins University, 1995 A Dissertation presented to the Graduate Faculty of the University of Virginia in Candidacy for the Degree of Doctor of Philosophy Department of Cell Biology University of Virginia May, 2004 Table of Contents Chapter 1 . 1 Chapter 2 . 48 Chapter 3 . 87 Chapter 4 . 123 Chapter 5 . 133 References . 137 Appendix A . 163 Appendix B . 209 Appendix C . 216 Appendix D . 223 Appendix E . 232 Appendix F . 234 Appendix G . 283 Appendix H . 318 Appendix I . 324 Abstract This research represents initial steps towards understanding the relation between changes in gene expression, mitochondrial function and cell death in cell-based models of Parkinson’s disease. The main hypothesis is that rapid gene expression changes in cells exposed to parkinsonian neurotoxins occur, are dependent on mitochondrial status, and directly impact intracellular signaling pathways that determine whether a cell lives or dies. Our cellular model is comprised of SH-SY5Y neuroblastoma cells exposed to the parkinsonian neurotoxin methylpyridinium ion. Transcriptomic changes are evaluated with nylon and glass-based cDNA microarray technology. Cardinal symptoms of Parkinson’s disease, characteristic pathological changes, therapeutic modalities, and current theories on the etiology of the disorder are discussed. Our results verify the existence of mitochondrial-nuclear signaling in the context of electron transport chain deficits, as well as suggesting the vital roles played in this process by previously described intracellular signaling pathways. These results will serve to direct future investigations into gene expression changes relevant to the processes of cell death and cell survival in our cellular model of Parkinson’s disease, and may provide important insights into the pathophysiology of the in vivo disease process. -

2006 Kyoto, Japan
October 28 - November 2, 2006 ~ Kyoto, Japan ~ Final Program Table of Contents Welcome . .2 Acknowledgements . .3 Organization . .6 MDS .Committees .& Task. .Forces . .9 International .Congress .Registration .and Venue. .12 International .Congress .Information . 13-15 . Continuing .Medical .Education . .13 . Evaluations . .14 . Press .Room . .15 Program-at-a-Glance . .17 Scientific Session Definitions . .19 Scientific .Sessions . .21 Faculty . .51 Committee .& Task. .Force .Meetings . .56 Exhibitor .Information . .57 Exhibitor .Directory . .58 Floor .Plans . 62-64 Map .of .Kyoto . .66 Lunch Map . .67 Subway Map . .68 Social Events . .69 Poster .Session .1 . .72 Poster .Session .2 . .88 Poster .Session .3 . .102 Poster .Session .4 . .117 CME .Request .Form . .133 The Movement. .Disorder .Society’s 0th International Congress of Parkinson’s Disease and Movement Disorders Welcome Letter Dear Colleagues, On behalf of The Movement Disorder Society (MDS), we are pleased to welcome you to Kyoto, Japan for the 10th International Congress of Parkinson’s Disease and Movement Disorders . The 10th International Congress has been designed to provide an innovative and comprehensive overview of the latest perspectives and research developments in the field of Movement Disorders . We encourage you to take every opportunity to participate in the Scientific Program which has drawn world renowned speakers and foremost experts in their respective fields . In the next days, the latest research regarding Movement Disorders will be presented and discussed in an open format, offering unique educational opportunities for all delegates . The International Congress convenes with a series of Opening Seminars and then continues with an array of Plenary, Parallel, Poster and Video Sessions, as well as Lunch Seminars, Controversies and Skills Workshops . -

Supplementary Table 3 Gene Microarray Analysis: PRL+E2 Vs
Supplementary Table 3 Gene microarray analysis: PRL+E2 vs. control ID1 Field1 ID Symbol Name M Fold P Value 69 15562 206115_at EGR3 early growth response 3 2,36 5,13 4,51E-06 56 41486 232231_at RUNX2 runt-related transcription factor 2 2,01 4,02 6,78E-07 41 36660 227404_s_at EGR1 early growth response 1 1,99 3,97 2,20E-04 396 54249 36711_at MAFF v-maf musculoaponeurotic fibrosarcoma oncogene homolog F 1,92 3,79 7,54E-04 (avian) 42 13670 204222_s_at GLIPR1 GLI pathogenesis-related 1 (glioma) 1,91 3,76 2,20E-04 65 11080 201631_s_at IER3 immediate early response 3 1,81 3,50 3,50E-06 101 36952 227697_at SOCS3 suppressor of cytokine signaling 3 1,76 3,38 4,71E-05 16 15514 206067_s_at WT1 Wilms tumor 1 1,74 3,34 1,87E-04 171 47873 238623_at NA NA 1,72 3,30 1,10E-04 600 14687 205239_at AREG amphiregulin (schwannoma-derived growth factor) 1,71 3,26 1,51E-03 256 36997 227742_at CLIC6 chloride intracellular channel 6 1,69 3,23 3,52E-04 14 15038 205590_at RASGRP1 RAS guanyl releasing protein 1 (calcium and DAG-regulated) 1,68 3,20 1,87E-04 55 33237 223961_s_at CISH cytokine inducible SH2-containing protein 1,67 3,19 6,49E-07 78 32152 222872_x_at OBFC2A oligonucleotide/oligosaccharide-binding fold containing 2A 1,66 3,15 1,23E-05 1969 32201 222921_s_at HEY2 hairy/enhancer-of-split related with YRPW motif 2 1,64 3,12 1,78E-02 122 13463 204015_s_at DUSP4 dual specificity phosphatase 4 1,61 3,06 5,97E-05 173 36466 227210_at NA NA 1,60 3,04 1,10E-04 117 40525 231270_at CA13 carbonic anhydrase XIII 1,59 3,02 5,62E-05 81 42339 233085_s_at OBFC2A oligonucleotide/oligosaccharide-binding -

Novel Targets of Apparently Idiopathic Male Infertility
International Journal of Molecular Sciences Review Molecular Biology of Spermatogenesis: Novel Targets of Apparently Idiopathic Male Infertility Rossella Cannarella * , Rosita A. Condorelli , Laura M. Mongioì, Sandro La Vignera * and Aldo E. Calogero Department of Clinical and Experimental Medicine, University of Catania, 95123 Catania, Italy; [email protected] (R.A.C.); [email protected] (L.M.M.); [email protected] (A.E.C.) * Correspondence: [email protected] (R.C.); [email protected] (S.L.V.) Received: 8 February 2020; Accepted: 2 March 2020; Published: 3 March 2020 Abstract: Male infertility affects half of infertile couples and, currently, a relevant percentage of cases of male infertility is considered as idiopathic. Although the male contribution to human fertilization has traditionally been restricted to sperm DNA, current evidence suggest that a relevant number of sperm transcripts and proteins are involved in acrosome reactions, sperm-oocyte fusion and, once released into the oocyte, embryo growth and development. The aim of this review is to provide updated and comprehensive insight into the molecular biology of spermatogenesis, including evidence on spermatogenetic failure and underlining the role of the sperm-carried molecular factors involved in oocyte fertilization and embryo growth. This represents the first step in the identification of new possible diagnostic and, possibly, therapeutic markers in the field of apparently idiopathic male infertility. Keywords: spermatogenetic failure; embryo growth; male infertility; spermatogenesis; recurrent pregnancy loss; sperm proteome; DNA fragmentation; sperm transcriptome 1. Introduction Infertility is a widespread condition in industrialized countries, affecting up to 15% of couples of childbearing age [1]. It is defined as the inability to achieve conception after 1–2 years of unprotected sexual intercourse [2]. -

Aes Corporation
THE AES CORPORATION THE AES CORPORATION The global power company A Passion to Serve A Passion A PASSION to SERVE 2000 ANNUAL REPORT ANNUAL REPORT THE AES CORPORATION 1001 North 19th Street 2000 Arlington, Virginia 22209 USA (703) 522-1315 CONTENTS OFFICES 1 AES at a Glance AES CORPORATION AES HORIZONS THINK AES (CORPORATE OFFICE) Richmond, United Kingdom Arlington, Virginia 2 Note from the Chairman 1001 North 19th Street AES OASIS AES TRANSPOWER Arlington, Virginia 22209 Suite 802, 8th Floor #16-05 Six Battery Road 5 Our Annual Letter USA City Tower 2 049909 Singapore Phone: (703) 522-1315 Sheikh Zayed Road Phone: 65-533-0515 17 AES Worldwide Overview Fax: (703) 528-4510 P.O. Box 62843 Fax: 65-535-7287 AES AMERICAS Dubai, United Arab Emirates 33 AES People Arlington, Virginia Phone: 97-14-332-9699 REGISTRAR AND Fax: 97-14-332-6787 TRANSFER AGENT: 83 2000 AES Financial Review AES ANDES FIRST CHICAGO TRUST AES ORIENT Avenida del Libertador COMPANY OF NEW YORK, 26/F. Entertainment Building 602 13th Floor A DIVISION OF EQUISERVE 30 Queen’s Road Central 1001 Capital Federal P.O. Box 2500 Hong Kong Buenos Aires, Argentina Jersey City, New Jersey 07303 Phone: 852-2842-5111 Phone: 54-11-4816-1502 USA Fax: 852-2530-1673 Fax: 54-11-4816-6605 Shareholder Relations AES AURORA AES PACIFIC Phone: (800) 519-3111 100 Pine Street Arlington, Virginia STOCK LISTING: Suite 3300 NYSE Symbol: AES AES ENTERPRISE San Francisco, California 94111 Investor Relations Contact: Arlington, Virginia USA $217 $31 Kenneth R. Woodcock 93% 92% AES ELECTRIC Phone: (415) 395-7899 $1.46* 91% Senior Vice President 89% Burleigh House Fax: (415) 395-7891 88% 1001 North 19th Street $.96* 18 Parkshot $.84* AES SÃO PAULO Arlington, Virginia 22209 Richmond TW9 2RG $21 Av.