Determination of the Geographical Origin of Walnuts (Juglans Regia L.) Using Near-Infrared Spectroscopy † and Chemometrics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Essential Wholesale & Labs Carrier Oils Chart
Essential Wholesale & Labs Carrier Oils Chart This chart is based off of the virgin, unrefined versions of each carrier where applicable, depending on our website catalog. The information provided may vary depending on the carrier's source and processing and is meant for educational purposes only. Viscosity Absorbtion Comparible Subsitutions Carrier Oil/Butter Color (at room Odor Details/Attributes Rate (Based on Viscosity & Absorbotion Rate) temperature) Description: Stable vegetable butter with a neutral odor. High content of monounsaturated oleic acid and relatively high content of natural antioxidants. Offers good oxidative stability, excellent Almond Butter White to pale yellow Soft Solid Fat Neutral Odor Average cold weather stability, contains occlusive properties, and can act as a moistening agent. Aloe Butter, Illipe Butter Fatty Acid Compositon: Palmitic, Stearic, Oleic, and Linoleic Description: Made from Aloe Vera and Coconut Oil. Can be used as an emollient and contains antioxidant properties. It's high fluidiy gives it good spreadability, and it can quickly hydrate while Aloe Butter White Soft Semi-Solid Fat Neutral Odor Average being both cooling and soothing. Fatty Acid Almond Butter, Illipe Butter Compostion: Linoleic, Oleic, Palmitic, Stearic Description: Made from by combinging Aloe Vera Powder with quality soybean oil to create a Apricot Kernel Oil, Broccoli Seed Oil, Camellia Seed Oil, Evening Aloe Vera Oil Clear, off-white to yellow Free Flowing Liquid Oil Mild musky odor Fast soothing and nourishing carrier oil. Fatty Acid Primrose Oil, Grapeseed Oil, Meadowfoam Seed Oil, Safflower Compostion: Linoleic, Oleic, Palmitic, Stearic Oil, Strawberry Seed Oil Description: This oil is similar in weight to human sebum, making it extremely nouirshing to the skin. -
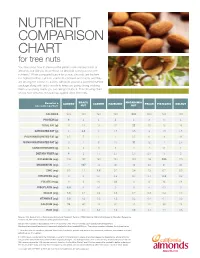
Nutrient Comparison Chart
NUTRIENT COMPARISON CHART for tree nuts You may know how to measure the perfect one-ounce portion of almonds, but did you know those 23 almonds come packed with nutrients? When compared ounce for ounce, almonds are the tree nut highest in fiber, calcium, vitamin E, riboflavin and niacin, and they are among the lowest in calories. Almonds provide a powerful nutrient package along with tasty crunch to keep you going strong, making them a satisfying snack you can feel good about. The following chart shows how almonds measure up against other tree nuts. BRAZIL MACADAMIA Based on a ALMOND CASHEW HAZELNUT PECAN PISTACHIO WALNUT one-ounce portion1 NUT NUT CALORIES 1602 190 160 180 200 200 160 190 PROTEIN (g) 6 4 4 4 2 3 6 4 TOTAL FAT (g) 14 19 13 17 22 20 13 19 SATURATED FAT (g) 1 4.5 3 1.5 3.5 2 1.5 1.5 POLYUNSATURATED FAT (g) 3.5 7 2 2 0.5 6 4 13 MONOUNSATURATED FAT (g) 9 7 8 13 17 12 7 2.5 CARBOHYDRATES (g) 6 3 9 5 4 4 8 4 DIETARY FIBER (g) 4 2 1.5 2.5 2.5 2.5 3 2 POTASSIUM (mg) 208 187 160 193 103 116 285 125 MAGNESIUM (mg) 77 107 74 46 33 34 31 45 ZINC (mg) 0.9 1.2 1.6 0.7 0.4 1.3 0.7 0.9 VITAMIN B6 (mg) 0 0 0.1 0.2 0.1 0.1 0.3 0.2 FOLATE (mcg) 12 6 20 32 3 6 14 28 RIBOFLAVIN (mg) 0.3 0 0.1 0 0 0 0.1 0 NIACIN (mg) 1.0 0.1 0.4 0.5 0.7 0.3 0.4 0.3 VITAMIN E (mg) 7.3 1.6 0.3 4.3 0.2 0.4 0.7 0.2 CALCIUM (mg) 76 45 13 32 20 20 30 28 IRON (mg) 1.1 0.7 1.7 1.3 0.8 0.7 1.1 0.8 Source: U.S. -

Is Peanut Butter Keto-Friendly?
Is Peanut Butter Keto-Friendly? Yes! Peanut butter contains carbohydrates, fat, and protein; the ratios of those macronutrients are what makes it acceptable for a keto diet. Our Once Again peanut butters that are made with one simple ingredient: peanuts, are a healthy addition to your keto diet. Peanuts are in the legume family, which includes a variety of beans. Generally speaking, legumes are not included in keto diets; however, peanuts have a higher fat content than other beans which makes them keto-friendly. Moreover, peanuts have a similar macronutrient distribution when compared to nuts such as almonds. They are high in fat and lower in carbs, and thus make a perfect food for the keto lifestyle. Peanut butter fits nicely into your keto diet as a dip for celery, or add it to smoothies, or experiment with it as an ingredient in salad dressings. Is Almond Butter Keto- Friendly? Yes! Almond butter is a staple for most following a keto lifestyle. Used often as a versatile ingredient in salad dressings, dips, and sauces, and in many other culinary innovations, nut butters in general have the desired macronutrient ration for a keto diet, and almond butter is a particularly good choice because of its fiber and vitamin E content. Most of our almond butters contain one simple ingredient: almonds. The newest addition to our product line is our Blanched Almond Butter, available in both in Extra Creamy and Crunchy varieties. Once Again Blanched Almond Butters contain only one gram of carbohydrate per serving! Blanching almonds involves removing the exterior of the almond, leaving behind a surprisingly nutty-sweet interior, with its lighter texture and color. -

Tea Seed Oil and Health Properties Fatih Seyis1, Emine
Tea Seed Oil and Health Properties Fatih Seyis1, Emine Yurteri1, Aysel Özcan1 1Recep Tayyip Erdoğan University: Faculty of Agriculture and Natural Science, Field Crops Department, Rize/Turkey, e-mail: [email protected] Abstract: Tea Oil has a mild fragrant flavor that goes with anything. It’s not a heavy oil like Olive Oil, but thinner – more like almond oil. If the taste or “oiliness” of olive oil overpowers your food. Along with its mild taste and pleasant tea-like aroma, this oil touts impressive health benefits. Tea seed oil has a high smoke point, contains more monounsaturated fatty acids than olive oil, contains fewer saturated fatty acids than olive oil, contains high levels of Vitamin E, polyphenol antioxidants and both Omegas 3 and 6, but has less Omega 6 and Polyunsaturated Fats than olive oil. Health Benefits of tea seed oil are: it can be applied topically and consumed internally to obtain its health benefits, camellia oil can be used for skin, hair, has anti-cancer effects, effects boost immunity and reduces oxidative stress. Camellia oil is used for a variety of other purposes, for example for cooking, as machinery lubricant, as ingredient in beauty products like night creams, salves, in hair care products and perfumes and is used to coat iron products to prevent rusting. Key words: Tea, seed oil, health 1. Introduction Like other genera of Camellia (from Theaceae family), the tea plant (C.sinensis) produces large oily seeds. In some countries where tea seed oil is abundantly available, it has been accepted as edible oil (Sahari et al., 2004). -

Bioactive Compounds in Nuts and Edible Seeds: Focusing on Brazil Nuts and Baru Almond of the Amazon and Cerrado Brazilian Biomes
Review Article SM Journal of Bioactive Compounds in Nuts and Nutrition and Edible Seeds: Focusing on Brazil Nuts Metabolism and Baru Almond of the Amazon and Cerrado Brazilian Biomes Egea MB1*, Lima DS1, Lodete AR1 and Takeuchi K1,2* 1Science and Technology, Goiano Institute of Education, Brazil 2Faculty of Nutrition, Federal University of Mato Grosso, Brazil Article Information Abstract Received date: Oct 09, 2017 The biodiversity of the Amazon and Cerrado biomes is extremely important for the populations that inhabit Accepted date: Nov 14, 2017 these areas, through the extractive collection of non-timber forest products such as fruits, nuts and edible seeds, which generate income and employment. Brazil nut (Bertholletia excelsa) is native from South America being Published date: Nov 20, 2017 found in the Amazon biome and baru almond (Dipteryx alata Vog.) is native from the Cerrado biome; these are part of the group of oleaginous that can be classified as true nuts and edible seeds, respectively. Both *Corresponding author are important sources of micronutrients that have been associated with several benefits to human health due to the presence of high levels of biologically active compounds such as minerals and vitamins. Minerals act Egea MB, Science and Technology, mostly as cofactors in various reactions, selenium has high availability in Brazil nuts and from selenocysteine Goiano Institute of Education, Brazil, and its enzymes, it exerts functions in the human body as an antioxidant, regulator of thyroid hormones and Tel: +55 64 36205636; protection of cardiovascular diseases. Among vitamins, tocopherol is a precursor to vitamin E, present in both Brazil nut and baru almond, being found in the form of α-tocopherol and having a role in the prevention of various Email: [email protected] diseases, including: cancer, diabetes, cataracts and cardiovascular and cerebrovascular diseases. -
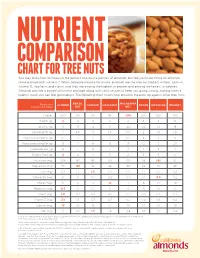
Chart for Tree Nuts
NUTRIENT COMPARISON CHART FOR TREE NUTS You may know how to measure the perfect one-ounce portion of almonds, but did you know those 23 almonds come packed with nutrients? When compared ounce for ounce, almonds are the tree nut highest in fiber, calcium, vitamin E, riboflavin and niacin, and they are among the highest in protein and among the lowest in calories. Almonds provide a powerful nutrient package along with tasty crunch to keep you going strong, making them a healthy snack you can feel good about. The following chart shows how almonds measure up against other tree nuts. BRAZIL MACADAMIA Based on a ALMOND CASHEW HAZELNUT PECAN PISTACHIO WALNUT one-ounce portion1 NUT NUT Calories 1602 190 160 180 200 200 160 190 Protein (g) 6 4 4 4 2 3 6 4 Total Fat (g) 14 19 13 17 22 20 13 18 Saturated Fat (g) 1 4.5 3 1.5 3.5 2 1.5 1.5 Polyunsaturated Fat (g) 3.5 7 2 2 0.5 6 4 13 Monounsaturated Fat (g) 9 7 8 13 17 12 7 2.5 Carbohydrates (g) 6 3 9 5 4 4 8 4 Dietary Fiber (g) 4 2 1 3 2 3 3 2 Potassium (mg) 208 187 160 193 103 116 285 125 Magnesium (mg) 77 107 74 46 33 34 31 45 Zinc (mg) 0.9 1.2 1.6 0.7 0.4 1.3 0.7 0.9 Vitamin B6 (mg) 0 0 0.1 0.2 0.1 0.1 0.3 0.2 Folate (mcg) 12 6 20 32 3 6 14 28 Riboflavin (mg) 0.3 0 0.1 0 0 0 0.1 0 Niacin (mg) 1.0 0.1 0.4 0.5 0.7 0.3 0.4 0.3 Vitamin E (mg) 7.3 1.6 0.3 4.3 0.2 0.4 0.6 0.2 Calcium (mg) 76 45 13 32 20 20 30 28 Iron (mg) 1.1 0.7 1.7 1.3 0.8 0.7 1.1 0.8 Source: U.S. -

Almond Cashew Barfi Recipe
150 Jackson St, Petone T: 568 4149 E: [email protected] ALMOND CASHEW BARFI RECIPE Almond Walnut Cashew Barfi is a healthy, quality substitute for candy that your entire family will enjoy Ingredients Makes 24 pcs 1/2 c almonds 1/2 c water 1/2 c walnuts 1/2 tsp cardamom powder 1/2 c cashew nuts 1 Tbsp sliced almonds to garnish 1 1/4 c sugar Method 1. Dry grind the walnuts, cashews and almonds in a food processor. 2. Dry roast the grounded nuts in a frying pan on low medium heat. 3. Roast them just enough so that the nuts start to give off an aroma. It will take about 4 to 5 minutes. Remove from heat and set aside. 4. Put the sugar and water together in a saucepan on medium heat. Bring to a boil to make the 1 thread syrup or on the candy thermometer it should reach 230 degrees F. 5. Turn off the heat and stir in the cardamom powder. 6. Add the nuts to the syrup and mix, and then spread over a greased 8-inch plate. Note: don’t let the syrup cool off. It must be spread while still hot. 7. Wait a few minutes until barfi is set but still soft. 8. Then cut the barfi into any shape you like (such as square, diamond, triangle). 9. Garnish each piece of barfi with sliced almonds while the barfi is still soft. 10. Allow the barfi to cool for about an hour to dry and hold its shape. -

Comparative Proteomic Analysis of Walnut (Juglans Regia L.) Pellicle Tissues Reveals the Regulation of Nut Quality Attributes
life Article Comparative Proteomic Analysis of Walnut (Juglans regia L.) Pellicle Tissues Reveals the Regulation of Nut Quality Attributes Paulo A. Zaini 1, Noah G. Feinberg 1, Filipa S. Grilo 2 , Houston J. Saxe 1 , Michelle R. Salemi 3, Brett S. Phinney 3 , Carlos H. Crisosto 1 and Abhaya M. Dandekar 1,* 1 Department of Plant Sciences, University of California, Davis, CA 95616, USA; [email protected] (P.A.Z.); [email protected] (N.G.F.); [email protected] (H.J.S.); [email protected] (C.H.C.) 2 Department of Food Sciences and Technology, University of California, Davis, CA 95616, USA; [email protected] 3 Proteomics Core Facility, University of California, Davis, CA 95616, USA; [email protected] (M.R.S.); [email protected] (B.S.P.) * Correspondence: [email protected] Received: 2 November 2020; Accepted: 25 November 2020; Published: 27 November 2020 Abstract: Walnuts (Juglans regia L.) are a valuable dietary source of polyphenols and lipids, with increasing worldwide consumption. California is a major producer, with ‘Chandler’ and ‘Tulare’ among the cultivars more widely grown. ‘Chandler’ produces kernels with extra light color at a higher frequency than other cultivars, gaining preference by growers and consumers. Here we performed a deep comparative proteome analysis of kernel pellicle tissue from these two valued genotypes at three harvest maturities, detecting a total of 4937 J. regia proteins. Late and early maturity stages were compared for each cultivar, revealing many developmental responses common or specific for each cultivar. Top protein biomarkers for each developmental stage were also selected based on larger fold-change differences and lower variance among replicates, including proteins for biosynthesis of lipids and phenols, defense-related proteins and desiccation stress-related proteins. -

Comparative Analysis of Mineral Elements and Essential Amino Acids Compositions in Juglans Sigillata and J
ZhaiAvailable M. et al./ online: Not Bot www.notulaebotanicae.ro Horti Agrobo, 2014, 42(1):36-42 Print ISSN 0255-965X; Electronic 1842-4309 Not Bot Horti Agrobo , 2014, 42(1):36-42 Comparative Analysis of Mineral Elements and Essential Amino Acids Compositions in Juglans sigillata and J. regia Walnuts Kernels Meizhi ZHAI*, Zhenyuan WANG, Dan WANG, Jing XU, Guanzhao SHI Northwest Agriculture & Forestry University, Walnut Research Center, College of Forestry, Yangling, Shaanxi, China; [email protected] (*corresponding author) Abstract Walnut kernel is famous for its high nutritional and economic values. The kernel is usually considered to be a good source of minerals and essential amino acids. In this paper, mineral elements (calcium, magnesium, iron, manganese, copper, and zinc) and essential amino acids (phenylalanine, valine, threonine, isoleucine, leucine, methionine, and lysine) compositions of kernels from 11 kinds of walnuts ( Juglans sigillata ) and 17 kinds of walnuts ( Juglans regia ) originated from China were determined by ICP-MS and HPLC, respectively. The order of nutritive mineral elements depending on their content (mg/100g) of samples was Mg> Ca> Zn> Mn> Fe> Cu in J. regia , while the order in J. sigillata was Mg> Ca> Mn> Fe > Zn > Cu. For essential amino acids, the order depending on the content (mg/g) of the essential amino acids in J. regia samples was leucine> isoleucine> valine> phenylalanine> lysine> threonine> methionine, while the order in J. sigillata was leucine> isoleucine> lysine> phenylalanine> valine> threonine> methionine. The kernels of walnuts ( J. regia and J. regia ) are good sources of health foods and dietary supplements. ‘Y029’ in Juglans sigillata and ‘XJ004’ in Juglans regia provided the best profiles of mineral elements and essential amino acids in comparison to others. -

View PDF of Aria
HAWAII OPERA THEATRE By Giuseppe Verdi February 1, 3 & 5, 2008 NEAL S. BLAISDELL CONCERT HALL oahu 2118 kalakaua avenue 808.921.1000, ala moana center 808.942.1148 maui the shops at wailea 808.879.1060 shop gucci.com Director’s Notes by HENRY G. AKINA Don Carlo is often considered Verdi’s most Wagnerian work. It makes use of the evolving structure of the Musiktheater, made so popular by Wagner, and introduces several motives that can be seen almost as Wagnerian leitmotifs. Verdi creates an entire political world set in Spain during the reign of Philip II. The opera follows four members of the Habsburg family: Philip II, King of all the world, son of an Emperor and ruler of a kingdom that “the sun never sets upon." Champion of the Catholic faith, devoted ruler and supporter of the Inquisition. This man rules his empire from a small room in the bowels of the Escurial palace. Likened by his enemies to a spider at the center of its web. At the time of the opera he has married three times. His current wife he married in spite of the fact that she had originally been promised to his son, Don Carlo. This marriage is one of the greatest burdens of his life. He does not know how to love. He is fiercely anti-protestant. His presence is felt oppressively in Flanders, a hot-bed of this new and troublesome movement. He has unleashed his armies and the Inquisition upon them. Elisabetta, Queen of Spain, daughter of Katharina de Medici and the King of France. -

Juglans Regiaregia Carpathiancarpathian Walnut,Walnut, Persianpersian Walnutwalnut
JuglansJuglans regiaregia CarpathianCarpathian walnut,walnut, PersianPersian walnutwalnut SEASONAL COLOURS jan feb mar apr mei jun jul aug sep okt nov dec TYPES OF PLANTING Tree types: standard trees, half-stem trees, multi-stemmed trees, trees for climbing, characteristic trees, fruit trees, woodland planting stock | Topiary on stem: vase, multi-stem umbrella USE Location: park, central reservation, roof garden, large garden, traffic areas, industrial zones, countryside, ecological zone | Pavement: none | Planting concepts: food forest, Landscape planting, Prairie planting CHARACTERISTICS Crown shape: flattened spherical | Crown structure: semi-open | Height: 20 - 30 m | Width: 20 - 30 m | Winter hardiness zone: 5B - 9B ASPECTS Wind: tolerant to wind | Soil: loess, sabulous clay, light clay, sand, loamy soil | Nutrient level: moderately rich in nutrients, rich in nutrients | Soil moisture level: dry, moist | Light requirements: sun | pH range: neutral, alkaline | Host plant/forage plant: birds, small mammals, humans | Extreme environments: tolerates air pollution PLANTKENMERKEN Flowers: catkins, pendulous | Flower colour: yellow-green | Flowering period: April - May | Leaf colour: buds bronze, green, underside pale green | Leaves: deciduous, ovoid, big, leathery , composite | Autumn colour: yellow-brown | Fruits: striking, edible, large, nut, drupe | Fruit colour: brown, green | Bark colour: pale grey | Bark: deeply furrowed, smooth, later on rough | Twig colour: pale grey | Twigs: bare | Root system: deep, fine roots, tap root Powered by TCPDF (www.tcpdf.org). -

Juglandaceae (Walnuts)
A start for archaeological Nutters: some edible nuts for archaeologists. By Dorian Q Fuller 24.10.2007 Institute of Archaeology, University College London A “nut” is an edible hard seed, which occurs as a single seed contained in a tough or fibrous pericarp or endocarp. But there are numerous kinds of “nuts” to do not behave according to this anatomical definition (see “nut-alikes” below). Only some major categories of nuts will be treated here, by taxonomic family, selected due to there ethnographic importance or archaeological visibility. Species lists below are not comprehensive but representative of the continental distribution of useful taxa. Nuts are seasonally abundant (autumn/post-monsoon) and readily storable. Some good starting points: E. A. Menninger (1977) Edible Nuts of the World. Horticultural Books, Stuart, Fl.; F. Reosengarten, Jr. (1984) The Book of Edible Nuts. Walker New York) Trapaceae (water chestnuts) Note on terminological confusion with “Chinese waterchestnuts” which are actually sedge rhizome tubers (Eleocharis dulcis) Trapa natans European water chestnut Trapa bispinosa East Asia, Neolithic China (Hemudu) Trapa bicornis Southeast Asia and South Asia Trapa japonica Japan, jomon sites Anacardiaceae Includes Piastchios, also mangos (South & Southeast Asia), cashews (South America), and numerous poisonous tropical nuts. Pistacia vera true pistachio of commerce Pistacia atlantica Euphorbiaceae This family includes castor oil plant (Ricinus communis), rubber (Hevea), cassava (Manihot esculenta), the emblic myrobalan fruit (of India & SE Asia), Phyllanthus emblica, and at least important nut groups: Aleurites spp. Candlenuts, food and candlenut oil (SE Asia, Pacific) Archaeological record: Late Pleistocene Timor, Early Holocene reports from New Guinea, New Ireland, Bismarcks; Spirit Cave, Thailand (Early Holocene) (Yen 1979; Latinis 2000) Rincinodendron rautanenii the mongongo nut, a Dobe !Kung staple (S.