Supplementary Exercises for Jalview 2.9 Alpha
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ACE-2019-Query-Builder-And-Tree
Copyright © 2019 by Aras Corporation. This material may be distributed only subject to the terms and conditions set forth in the Open Publication License, V1.0 or later (the latest version is presently available at http://www.opencontent.org/openpub/). Distribution of substantively modified versions of this document is prohibited without the explicit permission of the copyright holder. Distribution of the work or derivative of the work in any standard (paper) book form for a commercial purpose is prohibited unless prior permission is obtained from the copyright holder. Aras Innovator, Aras, and the Aras Corp "A" logo are registered trademarks of Aras Corporation in the United States and other countries. All other trademarks referenced herein are the property of their respective owners. Microsoft, Office, SQL Server, IIS and Windows are either registered trademarks or trademarks of Microsoft Corporation in the United States and/or other countries. Notice of Liability The information contained in this document is distributed on an "As Is" basis, without warranty of any kind, express or implied, including, but not limited to, the implied warranties of merchantability and fitness for a particular purpose or a warranty of non-infringement. Aras shall have no liability to any person or entity with respect to any loss or damage caused or alleged to be caused directly or indirectly by the information contained in this document or by the software or hardware products described herein. Copyright © 2019 by Aras Corporation. This material may be distributed only subject to the terms and conditions set forth in the Open Publication License, V1.0 or later (the latest version is presently available at http://www.opencontent.org/openpub/). -

Predicting Search Task Difficulty
Predicting Search Task Difficulty Jaime Arguello University of North Carolina at Chapel Hill [email protected] Abstract. Search task difficulty refers to a user's assessment about the amount of effort required to complete a search task. Our goal in this work is to learn predictive models of search task difficulty. We evaluate features derived from the user's interaction with the search engine as well as features derived from the user's level of interest in the task and level of prior knowledge in the task domain. In addition to user-interaction fea- tures used in prior work, we evaluate features generated from scroll and mouse-movement events on the SERP. In some situations, we may prefer a system that can predict search task difficulty early in the search session. To this end, we evaluate features in terms of whole-session evidence and first-round evidence, which excludes all interactions starting with the second query. Our results found that the most predictive features were different for whole-session vs. first-round prediction, that mouseover fea- tures were effective for first-round prediction, and that level of interest and prior knowledge features did not improve performance. 1 Introduction Search engine users engage in a wide variety of search tasks. A large body of prior research focused on characterizing different types of search tasks (see Li and Belkin [10]). The motivation behind this prior work is to understand how task characteristics influence search behavior and how search systems can provide customized interactions for different task types. One important search task characteristic is search task difficulty. -

Kurzweil 1000 Version 12 New Features
Kurzweil 1000 Version 12 New Features For the most up-to-date feature information, refer to the Readme file on the product CD. The following is a summary of what’s new in Version 12. For complete details, go to the online Manual by pressing Alt+H+O. Where applicable, Search key words are provided for you to use in the online Manual. • While you may not notice any difference, the internal structure of Kurzweil 1000 Version 12 has been overhauled and now uses Microsoft .NET Framework. The intent is to make it easier for Cambium Learning Technologies to develop features for the product going forward. • Note that Kurzweil 1000 Version 12 now supports 64-bit operating systems and Microsoft Windows 7 operating system. • As always, Kurzweil 1000 has the latest OCR engines, FineReader 9.0.1 and ScanSoft 16.2. The new ScanSoft version includes recognition languages from the Sami family. • An especially exciting new feature is the New User Wizard, a set of topics that introduces and walks new users through a number of Kurzweil 1000 features and preference setups. It appears when you start up Kurzweil 1000, but can be disabled and accessed from the Help menu by pressing Alt+H+W. (Search: New User Wizard.) • Currency Recognition has been updated to support new bills. Note that Currency Recognition now requires a color scanner. • New features and enhancements in reference tools include: 1. updates of the American Heritage Dictionary and Roget’s Thesaurus. 2. the ability to find up to 114 of your previously looked up entries; and last but not least the addition to dictionary and thesaurus lookup of human pronunciations and Anagrams. -

The Fancytooltips Package∗†
The fancytooltips package∗y Robert Maˇr´ık [email protected] May 4, 2009 1 Introduction The package fancytooltips is a package for LATEX. The pdf can be created by pdflatex or by latex + dvips + AdobeDistiller1 + Adobe Acrobat2. It allows to create tooltips in a similar way like cooltooltips package, but the tooltip is a page from another PDF file. In this way you can use mathematics, pictures and animations in your tooltips. The resulting PDF file can be used also with free Abobe Reader. The tooltips are activated by clicking the active area on the screen and deac- tivated after closing page or by moving mouse outside the link. You can try the links here (Einstein's formula) and also here (animation { numbers from 1 to 6). You have to use the free Adobe Reader or nonfree Adobe Acrobat to see the effect (xpdf, evince and others fail to work with JavaScripts). For more examples how the presentation may look like see the example.pdf and example-min.pdf files in the examples subdirectory. The buttons are created using eforms.sty which is a part of AcroTeX bundle. 2 Usage 2.1 The file with tooltips The file with tooltips is an ordinary pdf file, one tooltip per page, tooltips should be in the top right corner at the page, in a colored box and the rest of the page should be transparent. If you consider to use movetips option (see below), then every page should have the dimensions equal to the dimen- sions of the colored box with tooltip3. -

A Mobile Interface for Navigating Hierarchical Information Space$
Journal of Visual Languages and Computing 31 (2015) 48–69 Contents lists available at ScienceDirect Journal of Visual Languages and Computing journal homepage: www.elsevier.com/locate/jvlc A mobile interface for navigating hierarchical information space$ Abhishek P. Chhetri a,n, Kang Zhang b,c, Eakta Jain c,d a Computer Engineering Program, Erik Jonsson School of Engineering and Computer Science, University of Texas at Dallas, Richardson, TX 65080-3021, USA b School of Software Engineering, Tianjin University, Tianjin, China c Department of Computer Science, University of Texas at Dallas, Richardson, TX 65080-3021, USA d Texas Instruments, Dallas, TX, USA article info abstract Article history: This paper presents ERELT (Enhanced Radial Edgeless Tree), a tree visualization approach Received 2 June 2015 on modern mobile devices. ERELT is designed to offer a clear visualization of any tree Accepted 5 October 2015 structure with intuitive interaction. Such visualization can assist users in interacting with Available online 22 October 2015 a hierarchical structure such as a media collection, file system, etc. General terms: In the ERELT visualization, a subset of the tree is displayed at a time. The displayed tree Algorithms size depends on the maximum number of tree elements that can be put on the screen Design while maintaining clarity. Users can quickly navigate to the hidden parts of the tree Human factors through touch-based gestures. We have conducted a user study to evaluate this visuali- zation for a music collection. The study results show that this approach reduces the time Keywords: and effort in navigating tree structures for exploration and search tasks. -

Visualizing User Interface Events Event Stream Summarization Through Signs
Visualizing User Interface Events Event Stream Summarization through Signs Vagner Figueredo de Santana and Maria Cecília Calani Baranauskas Institute of Computing, University of Campinas (UNICAMP), Albert Einstein Av., 1251, Campinas, Brazil Keywords: Semiotics, User Interface Events, Visualization of Usage Data, Log Visualization, Event Icons, Icon Library, Image Library, User Interface Evaluation, Usability Evaluation, Accessibility Evaluation, Web Usage Mining. Abstract: Effective visual representation is related to how people interpret signs created to carry specific information. In the last years many user interface evaluation tools are considering detailed usage data to represent users’ actions. The volume of data gathered is leading developers to represent usage in a summarized way through graphical representations. If visual components used to represent complex data are not effective, then graphics used to summarize data may turn the interpretation of complex terms even harder. This work presents a study about graphical representations for user interface (UI) events and contributes with the validation of usage graph visualization and an open set of signs to support the summarization of client-side logs. The study involved 28 Information Technology specialists, potential users of UI evaluation tools. From the results one expects that evaluation tool developers, evaluators, and Web usage miners can reuse the validated usage graph representation and proposed set of signs to represent usage data in a summarized way. 1 INTRODUCTION these events is called, respectively, client-side logs and server-side logs (Santana and Baranauskas, The evaluation of user interface (UI) is a key task 2010a). when developing information systems and is part of In the last decade, website evaluation tools using a number of Software Engineering development server-side data (i.e., based on Web server logs) processes. -

Completeview™ Video Client User Manual
CompleteView™ Video Client User Manual CompleteView™ Version 4.7 Contents Introduction ................................................................................................................ 1 End User License Agreement ........................................................................................................................1 System Requirements ....................................................................................................................................3 Operation .................................................................................................................... 3 Getting Started ...............................................................................................................................................3 Starting the CompleteView Client ................................................................................................................3 Logging In to the CompleteView Client .......................................................................................................4 The Login Dialog .........................................................................................................................................4 Client Application Update ...........................................................................................................................5 Contacting Video Servers ............................................................................................................................6 Application Overview -

Clickjacking: Attacks and Defenses
Clickjacking: Attacks and Defenses Lin-Shung Huang Alex Moshchuk Helen J. Wang Carnegie Mellon University Microsoft Research Microsoft Research [email protected] [email protected] [email protected] Stuart Schechter Collin Jackson Microsoft Research Carnegie Mellon University [email protected] [email protected] Abstract such as a “claim your free iPad” button. Hence, when Clickjacking attacks are an emerging threat on the web. the user “claims” a free iPad, a story appears in the user’s In this paper, we design new clickjacking attack variants Facebook friends’ news feed stating that she “likes” the using existing techniques and demonstrate that existing attacker web site. For ease of exposition, our description clickjacking defenses are insufficient. Our attacks show will be in the context of web browsers. Nevertheless, the that clickjacking can cause severe damages, including concepts and techniques described are generally applica- compromising a user’s private webcam, email or other ble to all client operating systems where display is shared private data, and web surfing anonymity. by mutually distrusting principals. We observe the root cause of clickjacking is that an Several clickjacking defenses have been proposed and attacker application presents a sensitive UI element of a deployed for web browsers, but all have shortcomings. target application out of context to a user (such as hiding Today’s most widely deployed defenses rely on frame- the sensitive UI by making it transparent), and hence the busting [21, 37], which disallows a sensitive page from user is tricked to act out of context. To address this root being framed (i.e., embedded within another web page). -

Pen Computer Technology
Pen Computer Technology Educates the reader about the technologies involved in a pen computer Fujitsu PC Corporation www.fujitsupc.com For more information: [email protected] © 2002 Fujitsu PC Corporation. All rights reserved. This paper is intended to educate the reader about the technologies involved in a pen computer. After reading this paper, the reader should be better equipped to make intelligent purchasing decisions about pen computers. Types of Pen Computers In this white paper, "pen computer" refers to a portable computer that supports a pen as a user interface device, and whose LCD screen measures at least six inches diagonally. This product definition encompasses five generally recognized categories of standard products, listed in Table 1 below. PRODUCT TARGET PC USER STORAGE OPERATING RUNS LOCAL EXAMPLE CATEGORY MARKET INTERFACE SYSTEM PROGRAMS Webpad Consumer & No Standard Flash Windows CE, Only via Honeywell Enterprise browser memory Linux, QNX browser WebPAD II plug-ins CE Tablet Enterprise No Specialized Flash Windows CE Yes Fujitsu applications memory PenCentra Pen Tablet Enterprise Yes Windows & Hard drive Windows 9x, Yes Fujitsu specialized NT-4, 2000, Stylistic applications XP Pen-Enabled Consumer Yes Windows Hard drive Windows 9x, Yes Fujitsu & Enterprise 2000, XP LifeBook B Series Tablet PC Consumer Yes Windows Hard drive Windows XP Yes Many under & Enterprise Tablet PC development Edition Table 1: Categories of Pen Computers with LCD Displays of Six Inches or Larger Since the different types of pen computers are often confused, the following paragraphs are intended to help explain the key distinguishing characteristics of each product category. Pen Computers Contrasted Webpad: A Webpad's primary characteristic is that its only user interface is a Web browser. -

(12) Patent Application Publication (10) Pub. No.: US 2007/0211071 A1 Slotznick Et Al
US 20070211071A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2007/0211071 A1 Slotznick et al. (43) Pub. Date: Sep. 13, 2007 (54) METHOD AND APPARATUS FOR (52) U.S. Cl. .............................................................. 34.5/594 INTERACTING WITH A VISUALLY DISPLAYED DOCUMENT ON A SCREEN (57) ABSTRACT READER User interaction of a visually displayed document is pro vided via a graphical user interface (GUI). The document (76) Inventors: Benjamin Slotznick, Mt. Gretna, PA includes, and is parsed into, a plurality of text-based gram (US); Stephen C. Sheetz, Lititz, PA matical units. An input device modality is selected from a (US) plurality of input device modalities which determines the Correspondence Address: type of input device in which a user interacts with to make AKN GUMPSTRAUSS HAUER & FELD a selection. One or more grammatical units of the document L.L.P. are then selected using the selected type of input device. ONE COMMERCE SQUARE Each grammatical unit that is selected is read aloud to the 2005 MARKET STREET, SUITE 2200 user by loading the grammatical unit into a text-to-speech PHILADELPHIA, PA 19103 (US) engine. The text of the grammatical unit is thereby auto matically spoken. Furthermore, a Switching modality is (21) Appl. No.: 11/642,247 selected from a plurality of switching modalities. The (22) Filed: Dec. 20, 2006 Switching modality determines the manner in which one or more Switches are used to make a selection. Using the Related U.S. Application Data selected Switching modality, a user steps through at least Some of the grammatical units in an ordered manner by (60) Provisional application No. -

V9 User Manual (PDF)
This section of the manual gives a basic introduction to AB Tutor, and describes some of the concepts, such as network layout and passwords. AB Tutor manual Introduction What is AB Tutor? Basic ABT setup Advanced setup options Introduction to passwords Startup passwords Connection password Startup switches Introduction What is AB Tutor? AB Tutor is a networked classroom, instruction, monitoring and teaching tool that lets you train students in a networked classroom or lab, simply, effectively, at a very affordable price. Teachers, trainers and administrators can use the software to easily control, manage, monitor and support their students. This is a list of the key features of AB Tutor Cross platform Windows central server Windows and Mac tutor applications Windows and Mac client applications Computer Monitoring Real-time remote screen watch Simultaneous watch by multiple tutors Network efficient sizeable thumbnail views, with changeable refresh times Create different thumbnail arrangements for each group Monitor running applications and files Identify what site/file the student is working on Monitor and log student activity (applications, printing, websites and keystrokes) Take time and name-stamped snapshots of student activity Record and play back student screen activity Live search for users/computers Monitor multiple class groups simultaneously Keyword Notification Inform tutor when specific keywords are typed Automatically take snapshots of violations Trigger remote screen recording upon violation View and export all violations -
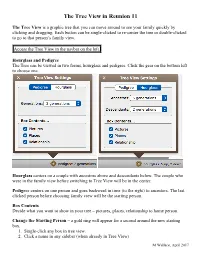
Tree View Handout
The Tree View in Reunion 11 The Tree View is a graphic tree that you can move around to see your family quickly by clicking and dragging. Each button can be single-clicked to re-center the tree or double-clicked to go to that person’s family view. Access the Tree View in the navbar on the left. Hourglass and Pedigree The Tree can be viewed in two forms, hourglass and pedigree. Click the gear on the bottom left to choose one. Hourglass centers on a couple with ancestors above and descendants below. The couple who were in the family view before switching to Tree View will be in the center. Pedigree centers on one person and goes backward in time (to the right) to ancestors. The last clicked person before choosing family view will be the starting person. Box Contents Decide what you want to show in your tree – pictures, places, relationship to home person. Change the Starting Person – a gold ring will appear for a second around the new starting box. 1. Single-click any box in tree view. 2. Click a name in any sidebar (when already in Tree View) M Wallace, April 2017 Page 2 of 2 3. Drag a person button from Family View to Tree View in navbar. 4. Drag a name from the sidebar to Tree View in navbar. 5. Control-click (right-click) any box in Tree View and choose someone from the Person menu. From this menu you can also - Mark/unmark the person - Mark everyone in this tree view - Switch back to Family View for that person - Open the Edit Person panel for that person - Show the Find Relative sidebar to re-calculate relationships for that person - Return to Starting Box – if you have moved around a lot and you want to go back to the beginning person/family, choose Edit > Locate Starting Box or Command B.