Sparklyr.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
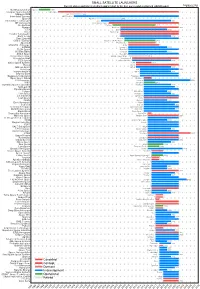
Small Satellite Launchers
SMALL SATELLITE LAUNCHERS NewSpace Index 2020/04/20 Current status and time from development start to the first successful or planned orbital launch NEWSPACE.IM Northrop Grumman Pegasus 1990 Scorpius Space Launch Demi-Sprite ? Makeyev OKB Shtil 1998 Interorbital Systems NEPTUNE N1 ? SpaceX Falcon 1e 2008 Interstellar Technologies Zero 2021 MT Aerospace MTA, WARR, Daneo ? Rocket Lab Electron 2017 Nammo North Star 2020 CTA VLM 2020 Acrux Montenegro ? Frontier Astronautics ? ? Earth to Sky ? 2021 Zero 2 Infinity Bloostar ? CASIC / ExPace Kuaizhou-1A (Fei Tian 1) 2017 SpaceLS Prometheus-1 ? MISHAAL Aerospace M-OV ? CONAE Tronador II 2020 TLON Space Aventura I ? Rocketcrafters Intrepid-1 2020 ARCA Space Haas 2CA ? Aerojet Rocketdyne SPARK / Super Strypi 2015 Generation Orbit GoLauncher 2 ? PLD Space Miura 5 (Arion 2) 2021 Swiss Space Systems SOAR 2018 Heliaq ALV-2 ? Gilmour Space Eris-S 2021 Roketsan UFS 2023 Independence-X DNLV 2021 Beyond Earth ? ? Bagaveev Corporation Bagaveev ? Open Space Orbital Neutrino I ? LIA Aerospace Procyon 2026 JAXA SS-520-4 2017 Swedish Space Corporation Rainbow 2021 SpinLaunch ? 2022 Pipeline2Space ? ? Perigee Blue Whale 2020 Link Space New Line 1 2021 Lin Industrial Taymyr-1A ? Leaf Space Primo ? Firefly 2020 Exos Aerospace Jaguar ? Cubecab Cab-3A 2022 Celestia Aerospace Space Arrow CM ? bluShift Aerospace Red Dwarf 2022 Black Arrow Black Arrow 2 ? Tranquility Aerospace Devon Two ? Masterra Space MINSAT-2000 2021 LEO Launcher & Logistics ? ? ISRO SSLV (PSLV Light) 2020 Wagner Industries Konshu ? VSAT ? ? VALT -

Redalyc.Status and Trends of Smallsats and Their Launch Vehicles
Journal of Aerospace Technology and Management ISSN: 1984-9648 [email protected] Instituto de Aeronáutica e Espaço Brasil Wekerle, Timo; Bezerra Pessoa Filho, José; Vergueiro Loures da Costa, Luís Eduardo; Gonzaga Trabasso, Luís Status and Trends of Smallsats and Their Launch Vehicles — An Up-to-date Review Journal of Aerospace Technology and Management, vol. 9, núm. 3, julio-septiembre, 2017, pp. 269-286 Instituto de Aeronáutica e Espaço São Paulo, Brasil Available in: http://www.redalyc.org/articulo.oa?id=309452133001 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative doi: 10.5028/jatm.v9i3.853 Status and Trends of Smallsats and Their Launch Vehicles — An Up-to-date Review Timo Wekerle1, José Bezerra Pessoa Filho2, Luís Eduardo Vergueiro Loures da Costa1, Luís Gonzaga Trabasso1 ABSTRACT: This paper presents an analysis of the scenario of small satellites and its correspondent launch vehicles. The INTRODUCTION miniaturization of electronics, together with reliability and performance increase as well as reduction of cost, have During the past 30 years, electronic devices have experienced allowed the use of commercials-off-the-shelf in the space industry, fostering the Smallsat use. An analysis of the enormous advancements in terms of performance, reliability and launched Smallsats during the last 20 years is accomplished lower prices. In the mid-80s, a USD 36 million supercomputer and the main factors for the Smallsat (r)evolution, outlined. -

Debris Mitigation, Assembly, Integration, and Test, in the Context of the Istsat-1 Project
Debris Mitigation, Assembly, Integration, and Test, in the context of the ISTsat-1 project Paulo Luís Granja Macedo Thesis to obtain the Master of Science Degree in Aerospace Engineering Supervisors: Prof. Paulo Jorge Soares Gil Prof. Agostinho Rui Alves da Fonseca Examination Committee Chairperson: Prof. José Fernando Alves da Silva Supervisor: Prof. Paulo Jorge Soares Gil Member of the Committee: Prof. Elsa Maria Pires Henriques November 2018 ii Dedicado ao meu Pai, Mae˜ e Irma˜ iii iv Acknowledgments I want to thank my supervisors, Professor Paulo Gil and Professor Agostinho Fonseca, for guiding me thorough the project. I also want to thank the ISTsat-1 team members, both professors and students, for giving me the opportunity to be part of such a great project and for the availability they had for my questions. The project would not happen if we did not have the University help and the ESA initiative Fly Your Satellite. I want to thank both organizations, that provided and will keep providing financial support, development rooms, test facilities and expertise in Space related matters. I want to thank all the other people that helped me through this phase, my fiends and girlfriend, thank you. Tambem´ quero agradecer aos meus pais e irma,˜ que me ajudaram em tudo o que foi preciso para chegar a este dia, sem eles nao˜ seria poss´ıvel. Obrigado. v vi Resumo O ISTsat-1 e´ um CubeSat desenvolvido por estudantes e professores do Instituto Superior Tecnico´ (IST), com a ajuda de um programa da ESA chamado Fly Your Satellite (FYS). O objectivo principal e´ educar estudantes em cienciaˆ e tecnologia espacial. -

Hawaii Space Flight Laboratory
Hawai`i Space Flight Laboratory University of Hawai`i at Mānoa Putting Rockets and Satellites into Orbit January 11, 2016 Director: Dr. Luke Flynn Contact Info: Email: [email protected] Phone: 808-956-3138 (Hawaii Space Grant) Web Site: http://www.hsfl.hawaii.edu Outline Why are Small Sats Relevant: Economics of Small Space Missions How did We Get There: Building to the ORS-4 Mission HSFL Mission Schedule and Future Plans Acknowledgment: This talk is given on behalf of the HSFL, HSGC, and HIGP staff who helped to make the ORS-4 Mission a possibility. Property of HSFL 2 Demand for “Space” In less than 60 years of space flight, the world has launched about 6500 satellites to space of which about 1000 are still operating… In the next 5 years, 3 companies (SpaceX/Google: 4000, OneWeb: 900, Samsung: 6000) will attempt to launch almost 11,000 small satellites….. They plan upgrades on 18-month cycles… Demand for space launch and small sats has shifted from Government to commercial groups. Property of HSFL 3 A View of the Launcher Market circa 2020 Market Launch Services Trend Market Size* Traditional Commercial Payload Type 2013-2020 2020 Primary Players Disruptors Large Satellites Flat $2.5B • Arianespace (Ariane 5) • SpaceX (FH) (> 4,000 kg) (20-25 sats) • ILS (Proton) • Commercial GEO Comm • Sea Launch (Zenit 3) • US Gov’t • ULA (Atlas V, Delta IV) Medium Satellites Flat $800M • Arianespace (Soyuz 2) • SpaceX (F9) (~ 1,500 – 4,000 kg) (~ 10 sats) • ILS (Angara-new) • Stratolauncher • Commercial GEO Comm • ULA (Atlas V, Delta II -

Aviation Catalog Av-14
® CHAMPION AEROSPACE LLC AVIATION CATALOG AV-14 REVISED JANUARY 2010 Spark Plugs Oil Filters Slick by Champion Exciters Leads Igniters ® Table of Contents SECTION PAGE Spark Plugs ........................................................................................................................................... 1 Product Features ....................................................................................................................................... 1 Spark Plug Type Designation System ............................................................................................................. 2 Spark Plug Types and Specifications ............................................................................................................. 3 Spark Plug by Popular Aircraft and Engines ................................................................................................ 4-12 Spark Plug Application by Engine Manufacturer .........................................................................................13-16 Other U. S. Aircraft and Piston Engines ....................................................................................................17-18 U. S. Helicopter and Piston Engines ........................................................................................................18-19 International Aircraft Using U. S. Piston Engines ........................................................................................ 19-22 Slick by Champion ............................................................................................................................. -

Atlas 5 Data Sheet
2/12/2018 Atlas 5 Data Sheet Space Launch Report: Atlas 5 Data Sheet Home On the Pad Space Logs Library Links Atlas 5 Vehicle Configurations Vehicle Components Atlas 5 Launch Record Atlas 5 was Lockheed Martin's Evolved Expendable Launch Vehicle (EELV) design for the U.S. Air Force. The United Launch Alliance consortium, a new company spun off by Boeing and Lockheed Martin, took over the Delta IV and Atlas V EELV programs in December 2006. The rocket, available in several variants, is built around a LOX/RP1 Common Core Booster (CCB) first stage and a LOX/LH2 Centaur second stage powered by one or two RL10 engines. Up to five solid rocket boosters (SRBs) can augment first stage thrust. A threedigit designator identifies Atlas V configurations. The first digit signifies the vehicle's payload fairing diameter in meters. The second digit tells the number of SRBs. The third digit provides the number of Centaur second stage RL10 engines (1 or 2). The Atlas V 400 series, with a 4 meter payload fairing and up to three SRBs, can boost up to 7.7 metric tons to a 28.7 deg geosynchronous transfer orbit (GTO) or 15.26 tonnes to a 28.7 deg low earth orbit (LEO) from Cape Canaveral. Atlas V 500, with a 5 meter diameter payload fairing and up to five SRBs, can put up to 8.9 tonnes to a 28.7 deg GTO or 18.85 tonnes into a 28.7 deg LEO. A 2.5 stage, Atlas 5 Heavy that uses three parallel burn CCBs, has been designed but not developed. -

Research Areas in Space
RESPOND & AI Capacity Building Programme Office ISRO HQ, Bengaluru RESPOND & AI Capacity Building Programme Office ISRO HQ, Bengaluru RESEARCH AREAS IN SPACE A Document for Preparing Research Project Proposals RESPOND & AI Capacity Building Programme Office ISRO HQ, Bengaluru January 2021 Technical Guidance Dr. M A Paul, Associate Director, RESPOND & AI, ISRO HQ Technical Support and Compilation Smt Nirupama Tiwari, Sci/Engr SE, CBPO, ISRO HQ Shri K Mahesh, Sr. Asst, CBPO, ISRO HQ Technical Guidance Dr. M A Paul, Associate Director, RESPOND & AI, ISRO HQ For any queries please contact Director, Capacity Building Programme Office (CBPO) Technical Support and Compilation Indian Space Research Organisation HQ Smt Nirupama Tiwari, Sci/Engr SE, CBPO, ISRO HQ Department of Space Shri K Mahesh, Sr. Asst, CBPO, ISRO HQ Government of India Antariksh Bhavan New BEL Road For any queries please contact Bangalore 560094 E-mail: [email protected] Director, Capacity Building Programme Office (CBPO) Indian Space Research Organisation HQ Department of Space Associate Director, RESPOND & Academic Interface Government of India Indian Space Research Organisation HQ Antariksh Bhavan Department of Space New BEL Road Government of India Bangalore 560094 Antariksh Bhavan E-mail: [email protected] New BEL Road Bangalore 560094 Associate Director, RESPOND & Academic Interface E-mail: [email protected] Indian Space Research Organisation HQ Department of Space Government of India Antariksh Bhavan New BEL Road Bangalore 560094 E-mail: [email protected] CONTENTS -

A SPARK Message Passing Framework for Cubesat Flight Software SPARK/Frama-C Conference - Copyright 2017 Carl Brandon & Peter Chapin Dr
CubedOS: A SPARK Message Passing Framework for CubeSat Flight Software SPARK/Frama-C Conference - Copyright 2017 Carl Brandon & Peter Chapin Dr. Carl Brandon & Dr. Peter Chapin [email protected] [email protected] Vermont Technical College +1-802-356-2822 (Brandon), +1-802-522-6763 (Chapin) 025226763 Randolph Center, VT 05061 USA http://www.cubesatlab.org Why We Use SPARK/Ada ELaNa IV lessons for CubeSat software: • NASA’s 2010 CubeSat Launch Initiative (ELaNa) • Our project was in the first group selected for launch • Our single-unit CubeSat was launched as part of NASA’s ELaNa IV on an Air Force ORS-3 Minotaur 1 flight November 19, 2013 to a 500 km altitude, 40.5o inclination orbit and remained in orbit until reentry over the central Pacific Ocean, November 21, 2016. Eight others were never heard from, two had partial contact for less than a week, and one worked for 4 months. • The Vermont Lunar CubeSat tested components of a Lunar navigation system in Low Earth Orbit Brandon & Chapin- SPARK/Frama-C Conference 2017 Vermont Lunar CubeSat It worked until our reentry on November 21, 2015: • We completed 11,071 orbits. • We travelled about 293,000,000 miles, equivalent to over 3/4 the distance to Jupiter. • Our single-unit CubeSat was launched as part of NASA’s ELaNa IV on an Air Force ORS-3 Minotaur 1 flight November 19, 2013 to a 500 km altitude, 40.5o inclination orbit and remained in orbit until November 21, 2016. It is the only one of the 12 ELaNa IV university CubeSats that operated until reentry, the last one quit 19 months earlier. -

Cubedos: a SPARK Message Passing Framework for Cubesat Flight Software NASA Academy of Aerospace Quality 2017 - Copyright 2017 Carl Brandon & Peter Chapin Dr
CubedOS: A SPARK Message Passing Framework for CubeSat Flight Software NASA Academy of Aerospace Quality 2017 - Copyright 2017 Carl Brandon & Peter Chapin Dr. Carl Brandon & Dr. Peter Chapin [email protected] [email protected] Vermont Technical College +1-802-356-2822 (Brandon), +1-802-522-6763 (Chapin) 025226763 Randolph Center, VT 05061 USA http://www.cubesatlab.org Why We Use SPARK/Ada ELaNa IV lessons for CubeSat software: • NASA’s 2010 CubeSat Launch Initiative (ELaNa) • Our project was in the first group selected for launch • Our single-unit CubeSat was launched as part of NASA’s ELaNa IV on an Air Force ORS-3 Minotaur 1 flight November 19, 2013 to a 500 km altitude, 40.5o inclination orbit and remained in orbit until reentry over the central Pacific Ocean, November 21, 2016, after two years and two days. Eight others were never heard from, two had partial contact for a few days, and one worked for 4 months. • The Vermont Lunar CubeSat tested components of a Lunar navigation system in Low Earth Orbit Brandon & Chapin- NASA Academy of Aerospace Quality 2017 Vermont Lunar CubeSat It worked until our reentry on November 21, 2015: • We completed 11,071 orbits. • We travelled about 293,000,000 miles, equivalent to over 3/4 the distance to Jupiter. • Our single-unit CubeSat was launched as part of NASA’s ELaNa IV on an Air Force ORS-3 Minotaur 1 flight November 19, 2013 to a 500 km altitude, 40.5o inclination orbit and remained in orbit until November 21, 2016. It is the only one of the 12 ELaNa IV university CubeSats that operated until reentry, the last one quit 19 months earlier. -

US Commercial Space Transportation Developments and Concepts
Federal Aviation Administration 2008 U.S. Commercial Space Transportation Developments and Concepts: Vehicles, Technologies, and Spaceports January 2008 HQ-08368.INDD 2008 U.S. Commercial Space Transportation Developments and Concepts About FAA/AST About the Office of Commercial Space Transportation The Federal Aviation Administration’s Office of Commercial Space Transportation (FAA/AST) licenses and regulates U.S. commercial space launch and reentry activity, as well as the operation of non-federal launch and reentry sites, as authorized by Executive Order 12465 and Title 49 United States Code, Subtitle IX, Chapter 701 (formerly the Commercial Space Launch Act). FAA/AST’s mission is to ensure public health and safety and the safety of property while protecting the national security and foreign policy interests of the United States during commercial launch and reentry operations. In addition, FAA/AST is directed to encourage, facilitate, and promote commercial space launches and reentries. Additional information concerning commercial space transportation can be found on FAA/AST’s web site at http://www.faa.gov/about/office_org/headquarters_offices/ast/. Federal Aviation Administration Office of Commercial Space Transportation i About FAA/AST 2008 U.S. Commercial Space Transportation Developments and Concepts NOTICE Use of trade names or names of manufacturers in this document does not constitute an official endorsement of such products or manufacturers, either expressed or implied, by the Federal Aviation Administration. ii Federal Aviation Administration Office of Commercial Space Transportation 2008 U.S. Commercial Space Transportation Developments and Concepts Contents Table of Contents Introduction . .1 Space Competitions . .1 Expendable Launch Vehicle Industry . .2 Reusable Launch Vehicle Industry . -

President's Message
Newsletter of the Amateur Radio Society of India (Indian Affiliate of IARU) October 2016 Issue President’s message ARSI welcomes The Mount Abu Amateur Radio organisation as its latest club Life Time Member As mentioned earlier, ARSI welcomes suggestions from all on how we can support and promote amateur radio activities- please write to the secretary and we will take up whatever is possible. 73, Gopal VU2GMN FROM THE EDITOR’S DESK A very well attended Wayanad Ham-meet was conducted over the weekend 24th / 25th September with 528 attendees! This was a first time event at Sultan Bathery, a small hill station in Kerala. The attendance would have been even more but for some road blockages due to an ongoing interstate dispute. Kudos to the organisers who spared no efforts We all are aware that Amateur Radio to make it a success. Communications play an important role I personally had to cancel my attendance at whenever there is a national calamity or the last moment and I really regret having emergency such as an Earthquake or Floods. done so! However, these days we hear about Amateur Radio being used during festivals, processions As usual ARSI keep working on pending issues and even during Elections in some states of with WPC. We have a new Wireless Advisor the country! Now – there are occasions when and so have to make a fresh start on dealing there is no other communications are with the issues. We understand that he will be available – with the cell phone towers down there for just a year. -

Design Study for a Formation-Flying Nanosatellite Cluster
2005:147 CIV EXAMENSARBETE Design Study for a Formation-Flying Nanosatellite Cluster SARA GIDLUND MASTER OF SCIENCE PROGRAMME in Space Engineering Luleå University of Technology Department of Space Science, Kiruna 2005:147 CIV • ISSN: 1402 - 1617 • ISRN: LTU - EX - - 05/147 - - SE Design Study for a Formation- Flying Nanosatellite Cluster. Centre for Large Space Structures and Systems Inc. (CLS3) 1425 Blvd. René Lévesque W-700 Montreal, Québec H3G 1T7 Canada Abstract Nanosatellites flying in formation vastly increase the capability of small satellite missions. The approach is within reach of technology, but there are severe challenges for the subsystems. The most demanding task is to implement effective control of the satellites. There are many options for solving the high and the low level control. The propulsion system and the communication system are the subsystems that will determine the constraints on the formation that may be used, and the duration of the mission. The mission of the cluster that the Centre for Large Space Structures and Systems Inc (CLS3) intends to launch will make use of existing technologies for nanosatellites and merge them with an in-house Guidance Navigation and Control system. The project will be developed as a collaboration between companies and organizations, in North America and Europe. The objective of the initial study was to summarize the state-of-the-art for the subsystems, and to stimulate discussions with potential partners. Negotiations with launch companies were also initiated. The satellites will fly in a circular or projected circular formation. In order to calculate the orbits of the satellites, it is of great importance to understand the Hill’s equations.