Modeling the Heat of Formation of Organic Compounds Using SPARC
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A-PDF Split DEMO : Purchase from to Remove the Watermark
A-PDF Split DEMO : Purchase from www.A-PDF.com to remove the watermark Figure 7.5 (a) Transition state for E2H-syn elimination. (b) Transition state for E2H-anti elimination. (c) Transition state for E,C elimination. tion state similar to that shown in Figure 7.5~is expected, since base attacks the molecule backside to the leaving group but frontside to the /3 hydrogen. Let us now turn to the experimental results to see if these predictions are borne out in fact. It has long been known that E2H reactions normally give preferentially anti elimination. For example, reaction of meso-stilbene dibromide with potassium ethoxide gives cis-bromostilbene (Reaction 7.39), whereas reaction of the D,L-dibromide gives the trans product (Reaction 7.40).lo2 A multitude of other examples exist-see, for example, note 64 (p. 355) and note 82 (p. 362). E2H reactions do, however, give syn elimination when: (1) an H-X di- hedral angle of 0" is achievable but one of 180" is not or, put another way, H and X can become syn-periplanar but not trans-periplanar ; (2) a syn hydrogen is much more reactive than the anti ones; (3) syn elimination is favored for steric reasons; and (4) an anionic base that remains coordinated with its cation, that, in turn, is coordinated with the leaving group, is used as catalyst. The very great importance of category 4 has only begun to be fully realized in the early 1970s. lo2 P. Pfeiffer, 2. Physik. Chem. Leibrig, 48, 40 (1904). 1,2-Elimination Reactions 371 An example of category 1 is found in the observation by Brown and Liu that eliminations from the rigid ring system 44, induced by the sodium salt of 2- cyclohexylcyclohexanol in triglyme, produces norborene (98 percent) but no 2-de~teronorbornene.~~~The dihedral angle between D and tosylate is O", but Crown ether present: No Yes that between H and tosylate is 120". -

Nomenclature Cyclic Aliphatic Hydrocarbons Are Named By
An alicyclic compound is an organic compound that is both aliphatic and cyclic. They contain one or more all-carbon rings which may be either saturated or unsaturated, but do not have aromatic character. Alicyclic compounds may have one or more aliphatic side chains attached. The simplest alicyclic compounds are the 1. monocyclic cycloalkanes: cyclopropane, cyclobutane, cyclopentane, cyclohexane, cyclohepta ne, cyclooctane, and so on. 2. Bicyclic alkanes include bicycloundecane, decalin, and housane. 3. Polycyclic alkanes include cubane, basketane, and tetrahedrane. Spiro compounds have two or more rings that are connected through only one carbon atom. Nomenclature Cyclic aliphatic hydrocarbons are named by prefixing cyclo- to the name of the corresponding open-chain hydrocarbon having the same number of carbons as the ring. For example: Cyclopropane Cyclobutane Cyclopentane Cyclopentene Substituents on the ring- alkyl, groups, halogens- are named and their positions indicated by numbers. Chlorocyclopropane 1,1- Dimethylyclopentane 1,3-Dimethylcyclohexane CH3 CH3 Cl H3C CH3 In simple cycloalkenes and cycloalkynes the double and triply bonded carbons are considered to occupy positions 1 and 2. For example: 3-Ethylcyclopentene 1,3-Cyclohexadiene H3C For convenience, aliphatic rings are often represented by simple geometric figures: a triangle for cyclopropane, a square for cyclobutane, a pentagon for cyclopentane, a hexagon for cyclohexane and so on. It is understood that two hydrogens are located at each corner of the figure unless some other group is indicated. For example H3C cyclopentane 3-Ethylcyclopentene 1,3-Cyclopentadiene CH3 CH3 Cl CH Cyclohexane 3 1,3-Dimethylcyclohexane 2- Chloro-1-methylcyclohexane As usual alcohols are given the ending –ol, which takes priority over –ene and appears last in the name. -

Principles of Surface Chemistry Central to the Reactivity of Organic Semiconductor Materials
Loyola University Chicago Loyola eCommons Dissertations Theses and Dissertations 2018 Principles of Surface Chemistry Central To the Reactivity of Organic Semiconductor Materials Gregory J. Deye Follow this and additional works at: https://ecommons.luc.edu/luc_diss Part of the Inorganic Chemistry Commons Recommended Citation Deye, Gregory J., "Principles of Surface Chemistry Central To the Reactivity of Organic Semiconductor Materials" (2018). Dissertations. 2952. https://ecommons.luc.edu/luc_diss/2952 This Dissertation is brought to you for free and open access by the Theses and Dissertations at Loyola eCommons. It has been accepted for inclusion in Dissertations by an authorized administrator of Loyola eCommons. For more information, please contact [email protected]. This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 License. Copyright © 2018 Gregory J Deye LOYOLA UNIVERSITY CHICAGO PRINCIPLES OF SURFACE CHEMISTRY CENTRAL TO THE REACTIVITY OF ORGANIC SEMICONDUCTOR MATERIALS A DISSERTATION SUBMITTED TO THE FACULTY OF THE GRADUATE SCHOOL IN CANDIDACY FOR THE DEGREE OF DOCTOR OF PHILOSOPHY PROGRAM IN CHEMISTRY BY GREGORY J. DEYE CHICAGO, IL AUGUST 2018 Copyright by Gregory J. Deye, 2018 All rights reserved. ACKNOWLEDGMENTS The scientific advances and scholarly achievements presented in this dissertation are a direct result of excellent mentorships, collaborations, and relationships for which I am very grateful. I thank my advisor, Dr. Jacob W. Ciszek, for his professionalism in mentorship and scientific discourse. He took special care in helping me give more effective presentations and seek elegant solutions to problems. It has been a privilege to work at Loyola University Chicago under his direction. I would also like to express gratitude to my committee members, Dr. -

New Applications of Cyclobutadiene Cycloadditions: Diversity and Target Oriented Synthesis
New Applications of Cyclobutadiene Cycloadditions: Diversity and Target Oriented Synthesis Author: Jason Joseph Marineau Persistent link: http://hdl.handle.net/2345/1741 This work is posted on eScholarship@BC, Boston College University Libraries. Boston College Electronic Thesis or Dissertation, 2010 Copyright is held by the author, with all rights reserved, unless otherwise noted. Boston College The Graduate School of Arts and Sciences Department of Chemistry NEW APPLICATIONS OF CYCLOBUTADIENE CYCLOADDITIONS: DIVERSITY AND TARGET ORIENTED SYNTHESIS a dissertation by JASON JOSEPH MARINEAU submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy December 2010 © Copyright by JASON JOSEPH MARINEAU 2010 New Applications of Cyclobutadiene Cycloadditions: Diversity and Target Oriented Synthesis Jason J. Marineau Thesis Advisor: Professor Marc. L. Snapper Abstract Cyclobutadiene cycloadditions provide rapid access to rigid polycyclic systems with high strain energy and unusual molecular geometries. Further functionalization of these systems allows entry into unexplored chemical space. A tricarbonylcyclobutadiene iron complex on solid support enables exploration of these cycloadditions in a parallel format amenable to diversity oriented synthesis. Modeling of the cycloaddition transition states with density functional calculations provides a theoretical basis for analysis of the regioselectivity observed in generation of these substituted bicyclo[2.2.0]hexene derivatives. The high strain energy accessible in cyclobutadiene cycloadducts and their derivatives renders them useful synthons for access to medium-ring natural products through ring expansion. Torilin, a guaiane sesquiterpene isolated from extracts of the fruits of Torilis japonica, exhibits a range of biological activities including testosterone 5 -reductase inhibition, hKv1.5 channel blocking, hepatoprotective, anti-inflammatory and anti-cancer effects. -

First Principles Prediction of Thermodynamic Properties
2 First Principles Prediction of Thermodynamic Properties Hélio F. Dos Santos and Wagner B. De Almeida NEQC: Núcleo de Estudos em Química Computacional, Departamento de Química, ICE Universidade Federal de Juiz de Fora (UFJF), Campus Universitário Martelos, Juiz de Fora LQC-MM: Laboratório de Química Computacional e Modelagem Molecular Departamento de Química, ICEx, Universidade Federal de Minas Gerais (UFMG) Campus Universitário, Pampulha, Belo Horizonte Brazil 1. Introduction The determination of the molecular structure is undoubtedly an important issue in chemistry. The knowledge of the tridimensional structure allows the understanding and prediction of the chemical-physics properties and the potential applications of the resulting material. Nevertheless, even for a pure substance, the structure and measured properties reflect the behavior of many distinct geometries (conformers) averaged by the Boltzmann distribution. In general, for flexible molecules, several conformers can be found and the analysis of the physical and chemical properties of these isomers is known as conformational analysis (Eliel, 1965). In most of the cases, the conformational processes are associated with small rotational barriers around single bonds, and this fact often leads to mixtures, in which many conformations may exist in equilibrium (Franklin & Feltkamp, 1965). Therefore, the determination of temperature-dependent conformational population is very much welcomed in conformational analysis studies carried out by both experimentalists and theoreticians. -

Here Science-Based Cost- Effective Pathways Forward?
Sponsors Institute of Atomic and Molecular Sciences, Academia Sinica, Taiwan PIRE-ECCI Program, UC Santa Barbara, USA Max-Planck-Gesellschaft, Germany National Science Council, Taiwan Organizing committee Dr. Kuei-Hsien Chen (IAMS, Academia Sinica, Taiwan) Dr. Susannah Scott (University of California - Santa Barbara, USA) Dr. Alec Wodtke (University of Göttingen & Max-Planck-Gesellschaft, Germany) Table of Contents General Information .............................................................................................. 1 Program for Sustainable Energy Workshop ....................................................... 3 I01 Dr. Alec M. Wodtke Beam-surface Scattering as a Probe of Chemical Reaction Dynamics at Interfaces ........................................................................................................... 7 I02 Dr. Kopin Liu Imaging the steric effects in polyatomic reactions ............................................. 9 I03 Dr. Chi-Kung Ni Energy Transfer of Highly Vibrationally Excited Molecules and Supercollisions ................................................................................................. 11 I04 Dr. Eckart Hasselbrink Energy Conversion from Catalytic Reactions to Hot Electrons in Thin Metal Heterostructures ............................................................................................... 13 I05 Dr. Jim Jr-Min Lin ClOOCl and Ozone Hole — A Catalytic Cycle that We Don’t Like ............... 15 I06 Dr. Trevor W. Hayton Nitric Oxide Reduction Mediated by a Nickel Complex -

Polycyclic Aromatic Hydrocarbon Structure Index
NIST Special Publication 922 Polycyclic Aromatic Hydrocarbon Structure Index Lane C. Sander and Stephen A. Wise Chemical Science and Technology Laboratory National Institute of Standards and Technology Gaithersburg, MD 20899-0001 December 1997 revised August 2020 U.S. Department of Commerce William M. Daley, Secretary Technology Administration Gary R. Bachula, Acting Under Secretary for Technology National Institute of Standards and Technology Raymond G. Kammer, Director Polycyclic Aromatic Hydrocarbon Structure Index Lane C. Sander and Stephen A. Wise Chemical Science and Technology Laboratory National Institute of Standards and Technology Gaithersburg, MD 20899 This tabulation is presented as an aid in the identification of the chemical structures of polycyclic aromatic hydrocarbons (PAHs). The Structure Index consists of two parts: (1) a cross index of named PAHs listed in alphabetical order, and (2) chemical structures including ring numbering, name(s), Chemical Abstract Service (CAS) Registry numbers, chemical formulas, molecular weights, and length-to-breadth ratios (L/B) and shape descriptors of PAHs listed in order of increasing molecular weight. Where possible, synonyms (including those employing alternate and/or obsolete naming conventions) have been included. Synonyms used in the Structure Index were compiled from a variety of sources including “Polynuclear Aromatic Hydrocarbons Nomenclature Guide,” by Loening, et al. [1], “Analytical Chemistry of Polycyclic Aromatic Compounds,” by Lee et al. [2], “Calculated Molecular Properties of Polycyclic Aromatic Hydrocarbons,” by Hites and Simonsick [3], “Handbook of Polycyclic Hydrocarbons,” by J. R. Dias [4], “The Ring Index,” by Patterson and Capell [5], “CAS 12th Collective Index,” [6] and “Aldrich Structure Index” [7]. In this publication the IUPAC preferred name is shown in large or bold type. -

Chem 22 Homework Set 12 1. Naphthalene Is Colorless, Tetracene
Chem 22 Homework set 12 1. Naphthalene is colorless, tetracene is orange, and azulene is blue. naphthalene tetracene azulene (a) Based on the colors observed for tetracene and azulene, what color or light does each compound absorb? (b) About what wavelength ranges do these colors correspond to? (c) Naphthalene has a conjugated π-system, so we know it must absorb somewhere in the UV- vis region of the EM spectrum. Where does it absorb? (d) What types of transitions are responsible for the absorptions? (e) Based on the absorption wavelengths, which cmpd has the smallest HOMO-LUMO gap? (f) How do you account for the difference in absorption λs of naphthalene vs tetracene? (g) Thinking about the factors that affect the absorption wavelengths, why does azulene not seem to follow the trend seen with the first two hydrocarbons? (h) Use the Rauk Hückelator (www.chem.ucalgary.ca/SHMO/) to determine the HOMO-LUMO gaps of each compound in β units. The use of this program will be demonstrated during Monday's class. 2. (a) What are the Hückel HOMO-LUMO gaps (in units of β) for the following molecules? Remember that we need to focus just on the π-systems. Use the Rauk Hückelator. (b) Use the Rauk Hückelator to draw some conjugated polyenes — linear as well as branched. Look at the HOMO. What is the correlation between the phases (ignore the sizes) of the p- orbitals that make up the HOMO and the positions of the double- and single-bonds in the Lewis structure? What is the relationship between the phases of p-orbitals of the LUMO to those of the HOMO? (c) Use your answer from part b and the pairing theorem to sketch the HOMO and LUMO of the polyenes below (again, just the phases — don't worry about the relative sizes of the p- orbitals). -
![Chemistry of Acenes, [60]Fullerenes, Cyclacenes and Carbon Nanotubes](https://docslib.b-cdn.net/cover/6902/chemistry-of-acenes-60-fullerenes-cyclacenes-and-carbon-nanotubes-516902.webp)
Chemistry of Acenes, [60]Fullerenes, Cyclacenes and Carbon Nanotubes
University of New Hampshire University of New Hampshire Scholars' Repository Doctoral Dissertations Student Scholarship Spring 2011 Chemistry of acenes, [60]fullerenes, cyclacenes and carbon nanotubes Chandrani Pramanik University of New Hampshire, Durham Follow this and additional works at: https://scholars.unh.edu/dissertation Recommended Citation Pramanik, Chandrani, "Chemistry of acenes, [60]fullerenes, cyclacenes and carbon nanotubes" (2011). Doctoral Dissertations. 574. https://scholars.unh.edu/dissertation/574 This Dissertation is brought to you for free and open access by the Student Scholarship at University of New Hampshire Scholars' Repository. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of University of New Hampshire Scholars' Repository. For more information, please contact [email protected]. CHEMISTRY OF ACENES, [60]FULLERENES, CYCLACENES AND CARBON NANOTUBES BY CHANDRANI PRAMANIK B.Sc., Jadavpur University, Kolkata, India, 2002 M.Sc, Indian Institute of Technology Kanpur, India, 2004 DISSERTATION Submitted to the University of New Hampshire in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in Materials Science May 2011 UMI Number: 3467368 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. UMI Dissertation Publishing UMI 3467368 Copyright 2011 by ProQuest LLC. All rights reserved. This edition of the work is protected against unauthorized copying under Title 17, United States Code. ProQuest LLC 789 East Eisenhower Parkway P.O. -

New Chemistry of Donor-Acceptor Cycloalkanes and Studies Towards the Synthesis of Streptorubin B
Western University Scholarship@Western Electronic Thesis and Dissertation Repository 8-2-2016 12:00 AM New Chemistry of Donor-Acceptor Cycloalkanes and Studies Towards the Synthesis of Streptorubin B Naresh Vemula The University of Western Ontario Supervisor Prof. Brian L. Pagenkopf The University of Western Ontario Graduate Program in Chemistry A thesis submitted in partial fulfillment of the equirr ements for the degree in Doctor of Philosophy © Naresh Vemula 2016 Follow this and additional works at: https://ir.lib.uwo.ca/etd Part of the Organic Chemistry Commons Recommended Citation Vemula, Naresh, "New Chemistry of Donor-Acceptor Cycloalkanes and Studies Towards the Synthesis of Streptorubin B" (2016). Electronic Thesis and Dissertation Repository. 3895. https://ir.lib.uwo.ca/etd/3895 This Dissertation/Thesis is brought to you for free and open access by Scholarship@Western. It has been accepted for inclusion in Electronic Thesis and Dissertation Repository by an authorized administrator of Scholarship@Western. For more information, please contact [email protected]. Abstract and Keywords Abstract This dissertation presents two separate chapters within the broad area of synthetic organic chemistry. The first chapter describes the annelation chemistry of donor-acceptor (DA) cyclopropanes and cyclobutanes for the synthesis of heterocycles. The Yb(OTf)3-catalyzed [4+2] cycloaddition between DA cyclobutanes and nitrosoarenes facilitated the synthesis of tetrahydro-1,2-oxazines in good to excellent yields as single diastereomers. Additionally, an unexpected deoxygenation occurred with electron-rich nitrosoarenes under MgI2-catalysis that afforded pyrrolidine products. The GaCl3-catalyzed [4+2] cycloaddition of DA cyclobutanes and cis-diazenes provided hexahydropyridazine derivatives in good to excellent yields as single diastereomers. -
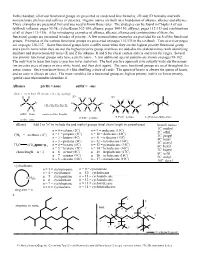
In This Handout, All of Our Functional Groups Are Presented As Condensed Line Formulas, 2D and 3D Formulas and with Nomenclature Prefixes and Suffixes (If Present)
In this handout, all of our functional groups are presented as condensed line formulas, 2D and 3D formulas and with nomenclature prefixes and suffixes (if present). Organic names are built on a foundation of alkanes, alkenes and alkynes. Those examples are presented first and you need to know those rules. The strategies can be found in Chapter 4 of our textbook (alkanes: pages 93-98, cycloalkanes 102-104, alkenes: pages 104-110, alkynes: pages 112-113 and combinations of all of them 113-115). After introducing examples of alkanes, alkenes, alkynes and combinations of them, the functional groups are presented in order of priority. A few nomenclature examples are provided for each of the functional groups. Examples of the various functional groups are presented on pages 115-135 in the textbook. Two overview pages are on pages 136-137. Some functional groups have a suffix name when they are the highest priority functional group and a prefix name when they are not the highest priority group, and these are added to the skeletal names with identifying numbers and stereochemistry terms (E and Z for alkenes, R and S for chiral centers and cis and trans for rings). Several low priority functional groups only have a prefix name. A few additional special patterns are shown on pages 98-102. The only way to learn this topic is practice (over and over). The best practice approach is to actually write out the names (on an extra piece of paper or on a white board, and then do it again). The same functional groups are used throughout the entire course. -

The X1 Method for Accurate and Efficient Prediction of Heats of Formation
THE JOURNAL OF CHEMICAL PHYSICS 127, 214105 ͑2007͒ The X1 method for accurate and efficient prediction of heats of formation ͒ Jianming Wu and Xin Xua State Key Laboratory of Physical Chemistry of Solid Surfaces and Center for Theoretical Chemistry, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, China ͑Received 31 July 2007; accepted 25 September 2007; published online 5 December 2007͒ We propose the X1 method which combines the density functional theory method with a neural network ͑NN͒ correction for an accurate yet efficient prediction of heats of formation. It calculates the final energy by using B3LYP/6-311+G͑3df,2p͒ at the B3LYP/6-311+G͑d,p͒ optimized geometry to obtain the B3LYP standard heats of formation at 298 K with the unscaled zero-point energy and thermal corrections at the latter basis set. The NN parameters cover 15 elements of H, Li, Be, B, C, N, O, F, Na, Mg, Al, Si, P, S, and Cl. The performance of X1 is close to the Gn theories, giving a mean absolute deviation of 1.43 kcal/mol for the G3/99 set of 223 molecules up to 10 nonhydrogen atoms and 1.48 kcal/mol for the X1/07 set of 393 molecules up to 32 nonhydrogen atoms. © 2007 American Institute of Physics. ͓DOI: 10.1063/1.2800018͔ I. INTRODUCTION the B3LYP method to calculate heats of formation of neutral organic molecules. Their results are promising. Calibrated ͑⌬ ͒ The accurate prediction of heats of formation Hf is with their own compilation of 180 organic molecules for the one of the central topics in computational chemistry.