Developmentally Regulated H2av Buffering Via Dynamic Sequestration
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Kaiso Is a Genome-Wide Repressor of Transcription That Is Essential for Amphibian Development Alexey Ruzov1,2,3,*, Donncha S
Research article 6185 Kaiso is a genome-wide repressor of transcription that is essential for amphibian development Alexey Ruzov1,2,3,*, Donncha S. Dunican1,3,*, Anna Prokhortchouk2, Sari Pennings1, Irina Stancheva1, Egor Prokhortchouk2 and Richard R. Meehan1,3,† 1Department of Biomedical Sciences, The University of Edinburgh, Hugh Robson Building, George Square, Edinburgh EH8 9XD, UK 2Institute of Gene Biology, Russian Academy of Sciences, Vavilova 34/5, Moscow, 119334, Russian Federation 3Human Genetics Unit, MRC, Western General Hospital, Crewe Road, Edinburgh EH4 2XU, UK *These authors contributed equally to this work †Author for correspondence (e-mail: [email protected]) Accepted 28 October 2004 Development 131, 6185-6194 Published by The Company of Biologists 2004 doi:10.1242/dev.01549 Summary DNA methylation in animals is thought to repress expression occurs before the mid-blastula transition transcription via methyl-CpG specific binding proteins, (MBT). Subsequent phenotypes (developmental arrest which recruit enzymatic machinery promoting the and apoptosis) strongly resemble those observed for formation of inactive chromatin at targeted loci. Loss of hypomethylated embryos. Injection of wild-type human DNA methylation can result in the activation of normally kaiso mRNA can rescue the phenotype and associated gene silent genes during mouse and amphibian development. expression changes of xKaiso-depleted embryos. Our Paradoxically, global changes in gene expression have not results, including gene expression profiling, are consistent been observed in mice that are null for the methyl-CpG with an essential role for xKaiso as a global repressor of specific repressors MeCP2, MBD1 or MBD2. Here, we methylated genes during early vertebrate development. -

Runx1 Gene in Acute Myeloid Leukemia: Expression Level Signifcance and Impact on Clinical Features
Runx1 Gene in Acute Myeloid Leukemia: Expression Level Signicance and Impact on Clinical Features Fadwa Said Abdelazim Mohamed ( [email protected] ) Cairo University Roxan E Shak National Cancer Institute, Cairo University Naglaa M. Hassan National Cancer Institute, Cairo University Research article Keywords: Acute myeloid leukemia, RUNX1, mutation, gene expression, Egypt Posted Date: April 7th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-20926/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/20 Abstract Background Acute myeloid leukemia represents the highest percentage of all adult acute leukemia variants. Runt-related transcription factor1 (RUNX1), a transcription factor with a known tumor suppressor function was recently reported as a tumor promotor in AML. We investigated the role of RUNX1 geneexpression levelin Egyptian AML patients and delineateits clinical signicance. Methods This study recruited 91 AML patients that were recently diagnosed at our hospital with 14 healthy age- and sex-matched donors of bone marrow transplantation unit. We measured RUNX1 gene expression level using reverse transcription–quantitative polymerase chain reaction. Results We found that RUNX1 gene expression level was signicantly higher compared to the control group (p < 0.001). Patients with FLT3 mutations had the higher expression level of RUNX1 (p=0.023). The male patients expressed signicantly higher level of RUNX1 (p=0.046). Conclusion The RUNX1 gene expression may serve as a diagnostic marker in Egyptian AML patients. Its relation to FLT3 may give clue that patients carrying this mutation may benet fromnew treatments that target RUNX1 in the future. -

Stages of Embryonic Development of the Zebrafish
DEVELOPMENTAL DYNAMICS 2032553’10 (1995) Stages of Embryonic Development of the Zebrafish CHARLES B. KIMMEL, WILLIAM W. BALLARD, SETH R. KIMMEL, BONNIE ULLMANN, AND THOMAS F. SCHILLING Institute of Neuroscience, University of Oregon, Eugene, Oregon 97403-1254 (C.B.K., S.R.K., B.U., T.F.S.); Department of Biology, Dartmouth College, Hanover, NH 03755 (W.W.B.) ABSTRACT We describe a series of stages for Segmentation Period (10-24 h) 274 development of the embryo of the zebrafish, Danio (Brachydanio) rerio. We define seven broad peri- Pharyngula Period (24-48 h) 285 ods of embryogenesis-the zygote, cleavage, blas- Hatching Period (48-72 h) 298 tula, gastrula, segmentation, pharyngula, and hatching periods. These divisions highlight the Early Larval Period 303 changing spectrum of major developmental pro- Acknowledgments 303 cesses that occur during the first 3 days after fer- tilization, and we review some of what is known Glossary 303 about morphogenesis and other significant events that occur during each of the periods. Stages sub- References 309 divide the periods. Stages are named, not num- INTRODUCTION bered as in most other series, providing for flexi- A staging series is a tool that provides accuracy in bility and continued evolution of the staging series developmental studies. This is because different em- as we learn more about development in this spe- bryos, even together within a single clutch, develop at cies. The stages, and their names, are based on slightly different rates. We have seen asynchrony ap- morphological features, generally readily identi- pearing in the development of zebrafish, Danio fied by examination of the live embryo with the (Brachydanio) rerio, embryos fertilized simultaneously dissecting stereomicroscope. -

Competition Between Histone and Transcription Factor Binding Regulates the Onset of Transcription in Zebrafish Embryos
1 Competition between histone and transcription factor binding regulates the 2 onset of transcription in zebrafish embryos 3 4 Shai R. Joseph1, Máté Pálfy1, Lennart Hilbert1,2,3, Mukesh Kumar1, Jens Karschau3, 5 Vasily Zaburdaev2,3, Andrej Shevchenko1, and Nadine L. Vastenhouw1# 6 7 1Max Planck Institute of Molecular Cell Biology and Genetics, 2Center for Systems 8 Biology Dresden, Pfotenhauerstraße 108, D-01307 Dresden, Germany, 3Max Planck 9 Institute for the Physics of Complex Systems, Nöthnitzerstraße 38, D-01187 Dresden, 10 Germany #Corresponding author 11 12 Email: [email protected] 13 1 14 SUMMARY 15 Upon fertilization, the genome of animal embryos remains transcriptionally inactive until 16 the maternal-to-zygotic transition. At this time, the embryo takes control of its 17 development and transcription begins. How the onset of zygotic transcription is regulated 18 remains unclear. Here, we show that a dynamic competition for DNA binding between 19 nucleosome-forming histones and transcription factors regulates zebrafish genome 20 activation. Taking a quantitative approach, we found that the concentration of non-DNA 21 bound core histones sets the time for the onset of transcription. The reduction in nuclear 22 histone concentration that coincides with genome activation does not affect nucleosome 23 density on DNA, but allows transcription factors to compete successfully for DNA 24 binding. In agreement with this, transcription factor binding is sensitive to histone levels 25 and the concentration of transcription factors also affects the time of transcription. Our 26 results demonstrate that the relative levels of histones and transcription factors regulate 27 the onset of transcription in the embryo. -

Vertebrate Embryonic Cleavage Pattern Determination
Chapter 4 Vertebrate Embryonic Cleavage Pattern Determination Andrew Hasley, Shawn Chavez, Michael Danilchik, Martin Wühr, and Francisco Pelegri Abstract The pattern of the earliest cell divisions in a vertebrate embryo lays the groundwork for later developmental events such as gastrulation, organogenesis, and overall body plan establishment. Understanding these early cleavage patterns and the mechanisms that create them is thus crucial for the study of vertebrate develop- ment. This chapter describes the early cleavage stages for species representing ray- finned fish, amphibians, birds, reptiles, mammals, and proto-vertebrate ascidians and summarizes current understanding of the mechanisms that govern these pat- terns. The nearly universal influence of cell shape on orientation and positioning of spindles and cleavage furrows and the mechanisms that mediate this influence are discussed. We discuss in particular models of aster and spindle centering and orien- tation in large embryonic blastomeres that rely on asymmetric internal pulling forces generated by the cleavage furrow for the previous cell cycle. Also explored are mechanisms that integrate cell division given the limited supply of cellular building blocks in the egg and several-fold changes of cell size during early devel- opment, as well as cytoskeletal specializations specific to early blastomeres A. Hasley • F. Pelegri (*) Laboratory of Genetics, University of Wisconsin—Madison, Genetics/Biotech Addition, Room 2424, 425-G Henry Mall, Madison, WI 53706, USA e-mail: [email protected] S. Chavez Division of Reproductive & Developmental Sciences, Oregon National Primate Research Center, Department of Physiology & Pharmacology, Oregon Heath & Science University, 505 NW 185th Avenue, Beaverton, OR 97006, USA Division of Reproductive & Developmental Sciences, Oregon National Primate Research Center, Department of Obstetrics & Gynecology, Oregon Heath & Science University, 505 NW 185th Avenue, Beaverton, OR 97006, USA M. -
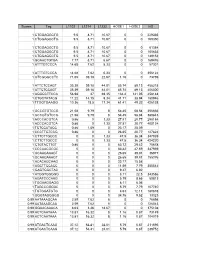
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108 -

(12) United States Patent (10) Patent No.: US 8.440,393 B2 Birrer Et Al
USOO8440393B2 (12) United States Patent (10) Patent No.: US 8.440,393 B2 Birrer et al. (45) Date of Patent: May 14, 2013 (54) PRO-ANGIOGENIC GENES IN OVARIAN OTHER PUBLICATIONS TUMORENDOTHELIAL CELL, SOLATES Boyd (The Basic Science of Oncology, 1992, McGraw-Hill, Inc., p. (75) Inventors: Michael J. Birrer, Mt. Airy, MD (US); 379). Tomas A. Bonome, Washington, DC Tockman et al. (Cancer Res., 1992, 52:2711s-2718s).* (US); Anil Sood, Pearland, TX (US); Pritzker (Clinical Chemistry, 2002, 48: 1147-1150).* Chunhua Lu, Missouri City, TX (US) Benedict et al. (J. Exp. Medicine, 2001, 193(1) 89-99).* Jiang et al. (J. Biol. Chem., 2003, 278(7) 4763-4769).* (73) Assignees: The United States of America as Matsushita et al. (FEBS Letters, 1999, vol. 443, pp. 348-352).* Represented by the Secretary of the Singh et al. (Glycobiology, 2001, vol. 11, pp. 587-592).* Department of Health and Human Abbosh et al. (Cancer Res. Jun. 1, 2006 66:5582-55.91 and Supple Services, Washington, DC (US); The mental Figs. S1-S7).* University of MD Anderson Cancer Zhai et al. (Chinese General Practice Aug. 2008, 11(8A): 1366 Center, Houston, TX (US) 1367).* Lu et al. (Cancer Res. Feb. 15, 2007, 64(4): 1757-1768).* (*) Notice: Subject to any disclaimer, the term of this Bagnato et al., “Activation of Mitogenic Signaling by Endothelin 1 in patent is extended or adjusted under 35 Ovarian Carcinoma Cells', Cancer Research, vol. 57, pp. 1306-1311, U.S.C. 154(b) by 194 days. 1997. Bouras et al., “Stanniocalcin 2 is an Estrogen-responsive Gene (21) Appl. -

Rapid Embryonic Cell Cycles Defer the Establishment of Heterochromatin by Eggless/Setdb1 in Drosophila
Downloaded from genesdev.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Rapid embryonic cell cycles defer the establishment of heterochromatin by Eggless/SetDB1 in Drosophila Charles A. Seller, Chun-Yi Cho, and Patrick H. O’Farrell Department of Biochemistry and Biophysics, University of California at San Francisco, San Francisco, California 94143, USA Acquisition of chromatin modifications during embryogenesis distinguishes different regions of an initially naïve genome. In many organisms, repetitive DNA is packaged into constitutive heterochromatin that is marked by di/ trimethylation of histone H3K9 and the associated protein HP1a. These modifications enforce the unique epigenetic properties of heterochromatin. However, in the early Drosophila melanogaster embryo, the heterochromatin lacks these modifications, which appear only later, when rapid embryonic cell cycles slow down at the midblastula transition (MBT). Here we focus on the initial steps restoring heterochromatic modifications in the embryo. We describe the JabbaTrap, a technique for inactivating maternally provided proteins in embryos. Using the JabbaTrap, we reveal a major requirement for the methyltransferase Eggless/SetDB1 in the establishment of heterochromatin. In contrast, other methyltransferases contribute minimally. Live imaging reveals that endogenous Eggless gradually accumulates on chromatin in interphase but then dissociates in mitosis, and its accumulation must restart in the next cell cycle. Cell cycle slowing as the embryo approaches the MBT permits increasing accumulation and action of Eggless at its targets. Experimental manipulation of interphase duration shows that cell cycle speed regulates Egg- less. We propose that developmental slowing of the cell cycle times embryonic heterochromatin formation. [Keywords: cell cycle; development; embryo; heterochromatin] Supplemental material is available for this article. -

1 Cytoplasmic Volume and Limiting Nucleoplasmin Scale Nuclear Size During Xenopus Laevis Development Pan Chen1, Miroslav Tomschi
bioRxiv preprint doi: https://doi.org/10.1101/511451; this version posted January 3, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Cytoplasmic volume and limiting nucleoplasmin scale nuclear size during Xenopus laevis development Pan Chen1, Miroslav Tomschik1, Katherine Nelson1,2, John Oakey2, J. C. Gatlin1, Daniel L. Levy1,* 1 Department of Molecular Biology, University of Wyoming, Laramie, WY, 82071 2 Department of Chemical Engineering, University of Wyoming, Laramie, WY, 82071 *Corresponding author and lead contact: Daniel L. Levy University of Wyoming Department of Molecular Biology 1000 E. University Avenue Laramie, WY, 82071 Phone: 307-766-4806 Fax: 307-766-5098 E-mail: [email protected] Running Head: Nucleoplasmin scales Xenopus nuclear size Abbreviations: NE, nuclear envelope; NPC, nuclear pore complex; ER, endoplasmic reticulum; NLS, nuclear localization signal; PKC, protein kinase C; CS, cross-sectional; N/C, nuclear-to-cytoplasmic; MBT, midblastula transition; Npm2, nucleoplasmin 1 bioRxiv preprint doi: https://doi.org/10.1101/511451; this version posted January 3, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. SUMMARY How nuclear size is regulated relative to cell size is a fundamental cell biological question. Reductions in both cell and nuclear sizes during Xenopus laevis embryogenesis provide a robust scaling system to study mechanisms of nuclear size regulation. -

An Optimized Streptavidin-Binding RNA Aptamer for Purification Of
Published online 23 October 2013 Nucleic Acids Research, 2014, Vol. 42, No. 2 e13 doi:10.1093/nar/gkt956 An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins Kathrin Leppek1,2,3 and Georg Stoecklin1,2,3,* 1Helmholtz Junior Research Group Posttranscriptional Control of Gene Expression, German Cancer Research Center (DKFZ), Im Neuenheimer Feld 280, 69120 Heidelberg, Germany, 2Zentrum fu¨ r Molekulare Biologie der Universita¨ t Heidelberg (ZMBH), Im Neuenheimer Feld 282, 69120 Heidelberg, Germany and 3DKFZ-ZMBH Downloaded from https://academic.oup.com/nar/article-abstract/42/2/e13/1030103 by guest on 11 March 2019 Alliance Received June 19, 2013; Revised September 6, 2013; Accepted September 27, 2013 ABSTRACT degradation (1,2). The fate of any given mRNA is essen- tially determined by the messenger ribonucleoprotein Determining the composition of messenger (mRNP) complex, i.e. the ensemble of proteins and regu- ribonucleoprotein (mRNP) particles is essential for latory RNAs an mRNA interacts with. Although decades a comprehensive understanding of the complex of research have uncovered specific functions of numerous mechanisms underlying mRNA regulation, but is RNA-binding proteins (RNA-BPs), surprisingly little is technically challenging. Here we present an RNA- known about the composition, heterogeneity and based method to identify RNP components using a dynamics of whole mRNPs. modified streptavidin (SA)-binding RNA aptamer There are essentially two strategies to explore the termed S1m. By optimizing the RNA aptamer S1 in content of mRNPs. On one hand, protein-based structure and repeat conformation, we improved its approaches rely on the purification of a particular RNA- affinity for SA and found a 4-fold repeat of S1m BP by immunoprecipitation together with its associated mRNAs, whose identity can then be determined by (4ÂS1m) to be more efficient than the established cDNA cloning, microarray analysis (RIP-Chip) or deep MS2 and PP7 systems from bacteriophages. -

Emergence of the Subapical Domain Is Associated with the Midblastula
bioRxiv preprint doi: https://doi.org/10.1101/713719; this version posted July 24, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Emergence of the subapical domain is associated with the midblastula 2 transition 3 Anja Schmidt (2), Jörg Großhans (1,2) 4 (1) Professur für Entwicklungsgenetik, Fachbereich Biologie, Philipps-Universität 5 Marburg, Karl-von-Frisch-Straße 8, 35043 Marburg, Germany 6 (2) Institut für Entwicklungsbiochemie, Universitätsmedizin, Georg-August- 7 Universität Göttingen, Justus-von-Liebig-Weg 11, 37077 Göttingen, Germany 8 Key words: cortical domains, epithelial domains, subapical, Canoe, midblastula 9 transition, zygotic genome activation 10 Abstract 11 Epithelial domains and cell polarity are determined by polarity proteins which are 12 associated with the cell cortex in a spatially restricted pattern. Early Drosophila 13 embryos are characterized by a stereotypic dynamic and de novo formation of cortical 14 domains. For example, the subapical domain emerges at the transition from syncytial 15 to cellular development during the first few minutes of interphase 14. The dynamics 16 in cortical patterning is revealed by the subapical markers Canoe/Afadin and ELMO- 17 Sponge, which widely distributed in interphase 13 but subapically restricted in 18 interphase 14. The factors and mechanism determining the timing for the emergence 19 of the subapical domain have been unknown. In this study, we show, that the restricted 20 localization of subapical markers depends on the onset of zygotic gene expression. -

Candidate Genes, Pathways and Mechanisms for Bipolar (Manic–Depressive) and Related Disorders: an Expanded Convergent Function
Molecular Psychiatry (2004), 1007–1029 & 2004 Nature Publishing Group All rights reserved 1359-4184/04 $30.00 www.nature.com/mp ORIGINAL RESEARCH ARTICLE Candidate genes, pathways and mechanisms for bipolar (manic–depressive) and related disorders: an expanded convergent functional genomics approach CA Ogden1,2,3, ME Rich1,2,3, NJ Schork2, MP Paulus2,3, MA Geyer2,3, JB Lohr2,3, R Kuczenski2,3 and AB Niculescu1,2,3 1Laboratory of Neurophenomics, University of California, San Diego, CA, USA; 2Department of Psychiatry, University of California, San Diego, CA, USA; 3VA San Diego Healthcare System VISN-22 MIRECC, San Diego, CA, USA Identifying genes for bipolar mood disorders through classic genetics has proven difficult. Here, we present a comprehensive convergent approach that translationally integrates brain gene expression data from a relevant pharmacogenomic mouse model (involving treatments with a stimulant—methamphetamine, and a mood stabilizer—valproate), with human data (linkage loci from human genetic studies, changes in postmortem brains from patients), as a bayesian strategy of crossvalidating findings. Topping the list of candidate genes, we have DARPP-32 (dopamine- and cAMP-regulated phosphoprotein of 32 kDa) located at 17q12, PENK (preproenkephalin) located at 8q12.1, and TAC1 (tachykinin 1, substance P) located at 7q21.3. These data suggest that more primitive molecular mechanisms involved in pleasure and pain may have been recruited by evolution to play a role in higher mental functions such as mood. The analysis also revealed other high-probability candidates genes (neurogenesis, neuro- trophic, neurotransmitter, signal transduction, circadian, synaptic, and myelin related), pathways and mechanisms of likely importance in pathophysiology.