Supplementary Table 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lack of Influence of Substrate on Ligand Interaction with the Human Multidrug
Downloaded from molpharm.aspetjournals.org at ASPET Journals on September 23, 2021 page 1 nd Interaction with the Human Multidrug Multidrug Human the with nd Interaction This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. -

TECHNISCHE UNIVERSITÄT MÜNCHEN Large Scale
TECHNISCHE UNIVERSITÄT MÜNCHEN Lehrstuhl für Genomorientierte Bioinformatik Large Scale Knowledge Extraction from Biomedical Literature Based on Semantic Role Labeling Thorsten Barnickel Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzende: Univ.‐Prof. Dr. I. Antes Prüfer der Dissertation: 1. Univ.‐Prof. Dr. H.‐W. Mewes 2. Univ.‐Prof. Dr. R. Zimmer (Ludwig‐Maximilians‐Universität München) Die Dissertation wurde am 30. Juli bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 25. November 2009 angenommen. ACKNOWLEDGEMENTS First and foremost, I would like to express my deep gratitude to my promoter Dr. Volker Stümpflen. Without his continuing, stimulating encouragement and his excellent background in Enterprise technologies still being predominantly used in the IT-industry rather than in academic research, I would not have been able to finish my doctorate in the presented form. Facing the tremendous amount of data that was generated by gathering the positional information of millions of biomedical terms, I was close to cutting the project down to a notably smaller version compared to the text mining system presented in this thesis. Volkers knowledge on database servers and performance tuning significantly contributed to the development of a database schema finally being able to cope with the immense amount of data. I would also like to cordially thank Prof. Dr. Hans-Werner Mewes, head of the Institute for Bioinformatics and Systems Biology (IBIS), for giving me the opportunity to do my doctorate at his institute and for his friendly support and encouragement all along my time at IBIS. -

Electronic Supplementary Material (ESI) for Molecular Biosystems
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is © The Royal Society of Chemistry 2015 Table S5 Mass data of proteins identified with label free shotgun proteomics a #Alt. Proteins; b Scores; c #Peptides; d SC [%]; e RMS90 [ppm]; f Rank; g Median(Controls:GLP1); h #(Controls:GLP1); i CV [%](Controls:GLP1); l Median(Controls:Palmitate); m #(Controls:Palmitate) ; n CV [%](Controls:Palmitate); o Median(Controls:GLP1 + Palmitate); p #(Controls:GLP1 + Palmitate); q CV [%](Controls:GLP1 + Palmitate) MW OK Accession Protein pI a b c d e f g h i l m n o p q [kDa] 78 kDa glucose-regulated protein OS=Rattus 1672.6 true GRP78_RAT 72.3 4.9 1 22 39 1.48 1 1.16 6 20.53 1.18 9 9.56 1.24 10 13.52 norvegicus GN=Hspa5 PE=1 SV=1 (M:1672.6) Endoplasmin OS=Rattus norvegicus 1253.0 true ENPL_RAT 92.7 4.6 1 23 29 3.34 2 0.85 8 33.34 1.19 6 39.58 1.18 10 28.11 GN=Hsp90b1 PE=1 SV=2 (M:1253.0) Stress-70 protein, mitochondrial OS=Rattus 1138.8 true GRP75_RAT 73.8 5.9 1 17 28.7 1.18 3 1.21 5 7.62 0.78 1 1.1 8 8.04 norvegicus GN=Hspa9 PE=1 SV=3 (M:1138.8) Protein disulfide-isomerase A3 OS=Rattus 1035.4 true PDIA3_RAT 56.6 5.8 1 16 35.2 2.84 4 0.94 3 17.22 1.15 4 26.62 1.25 4 7.86 norvegicus GN=Pdia3 PE=1 SV=2 (M:1035.4) Aconitate hydratase, mitochondrial 983.8 true ACON_RAT OS=Rattus norvegicus GN=Aco2 PE=1 85.4 8.7 1 19 31.4 2.72 5 0.89 2 23.6 1.21 2 9.25 0.89 3 32.87 (M:983.8) SV=2 60 kDa heat shock protein, mitochondrial 906.9 true CH60_RAT OS=Rattus norvegicus GN=Hspd1 PE=1 60.9 5.8 1 13 32.5 3.26 6 1.05 5 13.15 1.01 2 32.54 1.01 -

Taxifolin: a Wonder Molecule in Making Multiple Drug Targets
Short Communication Annals of Pharmacology and Pharmaceutics Published: 22 Aug, 2017 Taxifolin: A Wonder Molecule in Making Multiple Drug Targets Utkarsh Raj, Imlimaong Aier and Pritish Kumar Varadwaj* Department of Bioinformatics and Applied Sciences, Indian Institute of Information Technology, Allahabad, India Keywords Taxifolin; Anti-cancerous; Anti-ebola; Anti-oxidant; Anti-inflammatory Short Communication Taxifolin (often referred to as dihydroquercetin) is a flavanonol which is a part of the phytonutrient family (a set of chemical materials that occur evidently in flora and have more than one fitness benefits however aren't taken into consideration vital to human fitness). Taxifolin has been observed in a variety of cone bearing gymnosperms, like, Pinus roxburghii [1], Cedrus deodara [1], Larix sibirica (the Russian or Siberian larch) and inside the Chinese yew, Taxus chinensis var. mairei [2]. Its presence has also been reported in silymarin extract from the milk thistle seeds and cherry wood aged vinegar [3]. The 2D structure of Taxifolin has been depicted in Figure1. Over the past 50 years, almost six hundred studies (mostly conducted in Russia) have investigated the efficacy and protection of taxifolin. The important roles of the compound Taxifolin has been elucidated in Figure 2. The main highlights of this compound are its antioxidant efficiency and vascular-defensive movement. Taxifolin has also been shown to provide benefits to cardiovascular health, the pores and skin, cognitive feature, contamination, allergic reactions and immunodeficiency, in addition to the fitness of diabetics. Amongst others, research has examined that taxifolin is a brilliant antioxidant, far more effective than dietary carotenoids and it lowers blood viscosity and improves capillary microcirculation. -

Journal of the Hellenic Veterinary Medical Society
Journal of the Hellenic Veterinary Medical Society Vol. 71, 2020 Isobolographic analysis of analgesic interactions of silymarin with ketamine in mice NASER A.S. Department of physiology, biochemistry and pharmacology, Faculty of Veterinary Medicine, University of Mosul, Mosul, Iraq ALBADRANY Y. Department of physiology, biochemistry and pharmacology, Faculty of Veterinary Medicine, University of Mosul, Mosul, Iraq SHAABAN K.A. [email protected] https://doi.org/10.12681/jhvms.23653 Copyright © 2020 A.S. NASER, Y. ALBADRANY, K.A. SHAABAN To cite this article: NASER, A.S., ALBADRANY, Y., & SHAABAN, K.A. (2020). Isobolographic analysis of analgesic interactions of silymarin with ketamine in mice. Journal of the Hellenic Veterinary Medical Society, 71(2), 2171-2178. doi:https://doi.org/10.12681/jhvms.23653 http://epublishing.ekt.gr | e-Publisher: EKT | Downloaded at 23/09/2021 18:43:31 | J HELLENIC VET MED SOC 2020, 71(2): 2171-2178 Research article ΠΕΚΕ 2020, 71(2): 2171-2178 Ερευνητικό άρθρο Isobolographic analysis of analgesic interactions of silymarin with ketamine in mice A.S. Naser1*, Y. Albadrany2, K.A. Shaaban2 1Department of physiology, biochemistry and pharmacology, Faculty of Veterinary Medicine, University of Mosul, Mosul, Iraq 2Department of physiology, biochemistry and pharmacology, Faculty of Veterinary Medicine, University of Mosul, Mosul, Iraq ABSTRACT: The present study was undertaken to explore the analgesic effect of silymarin and ketamine alone or in combination in mice. Analgesia was measured by using a hot plate and the writhing test. The up-and-down method was used to determine the median effective analgesic dosages (ED50s) of silymarin and ketamine administered intraper- itoneally (ip) either alone or together. -

Epicatechin and Taxifolin Relevant for the Treatment of Hypertension and Viral Infection: Knowledge from Preclinical Studies
antioxidants Review Mechanisms Modified by (−)-Epicatechin and Taxifolin Relevant for the Treatment of Hypertension and Viral Infection: Knowledge from Preclinical Studies Iveta Bernatova 1,* and Silvia Liskova 1,2 1 Centre of Experimental Medicine, Institute of Normal and Pathological Physiology, Slovak Academy of Sciences, Sienkiewiczova 1, 813 71 Bratislava, Slovakia; [email protected] 2 Faculty of Medicine, Institute of Pharmacology and Clinical Pharmacology, Comenius University, Sasinkova 4, 811 08 Bratislava, Slovakia * Correspondence: [email protected]; Tel.: +421-2-32296013 Abstract: Various studies have shown that certain flavonoids, flavonoid-containing plant extracts, and foods can improve human health. Experimental studies showed that flavonoids have the capacity to alter physiological processes as well as cellular and molecular mechanisms associated with their antioxidant properties. An important function of flavonoids was determined in the cardiovascular system, namely their capacity to lower blood pressure and to improve endothelial function. (−)- Epicatechin and taxifolin are two flavonoids with notable antihypertensive effects and multiple beneficial actions in the cardiovascular system, but they also possess antiviral effects, which may be of particular importance in the ongoing pandemic situation. Thus, this review is focused on the Citation: Bernatova, I.; Liskova, S. current knowledge of (−)-epicatechin as well as (+)-taxifolin and/or (−)-taxifolin-modified biological Mechanisms Modified by action and underlining molecular mechanisms determined in preclinical studies, which are relevant − ( )-Epicatechin and Taxifolin not only to the treatment of hypertension per se but may provide additional antiviral benefits that Relevant for the Treatment of could be relevant to the treatment of hypertensive subjects with SARS-CoV-2 infection. Hypertension and Viral Infection: Knowledge from Preclinical Studies. -

Assembling the Puzzle of Taxifolin Polymorphism
molecules Article Assembling the Puzzle of Taxifolin Polymorphism Roman P. Terekhov 1,* , Irina A. Selivanova 1, Nonna A. Tyukavkina 1, Igor R. Ilyasov 1, Anastasiya K. Zhevlakova 1, Alexander V. Dzuban 2, Anatoliy G. Bogdanov 3, Georgiy N. Davidovich 3, Gennadii V. Shylov 4, Andrey N. Utenishev 1,4, Dmitriy Yu. Kovalev 5 , Anatoliy A. Fenin 6 and Tatyana G. Kabluchko 7 1 Department of Chemistry, Sechenov First Moscow State Medical University, Trubetskaya st. 8-2, 119991 Moscow, Russia; [email protected] (I.A.S.); [email protected] (N.A.T.); [email protected] (I.R.I.); [email protected] (A.K.Z.); [email protected] (A.N.U.) 2 Department of Chemistry, Lomonosov Moscow State University, Leninskiye Gory 1-3, 119991 Moscow, Russia; [email protected] 3 Faculty of Biology, Lomonosov Moscow State University, Leninskiye Gory 1-32, 119991 Moscow, Russia; [email protected] (A.G.B.); [email protected] (G.N.D.) 4 Laboratory of Structural Chemistry, Institute of Problems of Chemical Physics, Russian Academy of Sciences, Acad. Semenov av. 1, 143432 Chernogolovka, Russia; [email protected] 5 Laboratory of X-ray Investigation, Merzhanov Institute of Structural Macrokinetics and Materials Science, Russian Academy of Sciences, Acad. Osipyan str. 8, 142432 Chernogolovka, Russia; [email protected] 6 Institute of Materials for Modern Power Engineering and Nanotechnology, Mendeleev University of Chemical Technology of Russia, Miusskaya sq. 9, 125947 Moscow, Russia; [email protected] 7 Department of Technology, Ametis JSC, Naberezhnaya st. 68, 675000 Blagoveshchensk, Russia; [email protected] * Correspondence: terekhov_r_p@staff.sechenov.ru; Tel.: +7-965-232-5122 Academic Editor: Carlos Eduardo Sabino Bernardes Received: 15 October 2020; Accepted: 16 November 2020; Published: 20 November 2020 Abstract: A large amount of the current literature dedicated to solid states of active pharmaceutical ingredients (APIs) pays special attention to polymorphism of flavonoids. -

Murine Neonatal Ketogenesis Preserves Mitochondrial Energetics by Preventing Protein Hyperacetylation
ARTICLES https://doi.org/10.1038/s42255-021-00342-6 Murine neonatal ketogenesis preserves mitochondrial energetics by preventing protein hyperacetylation Yuichiro Arima 1,2,13 ✉ , Yoshiko Nakagawa3,13, Toru Takeo 3,13, Toshifumi Ishida 1, Toshihiro Yamada1, Shinjiro Hino4, Mitsuyoshi Nakao4, Sanshiro Hanada 2, Terumasa Umemoto 2, Toshio Suda2, Tetsushi Sakuma 5, Takashi Yamamoto5, Takehisa Watanabe6, Katsuya Nagaoka6, Yasuhito Tanaka6, Yumiko K. Kawamura7,8, Kazuo Tonami7, Hiroki Kurihara7, Yoshifumi Sato9, Kazuya Yamagata9,10, Taishi Nakamura 1,11, Satoshi Araki1, Eiichiro Yamamoto1, Yasuhiro Izumiya1,12, Kenji Sakamoto1, Koichi Kaikita1, Kenichi Matsushita 1, Koichi Nishiyama2, Naomi Nakagata3 and Kenichi Tsujita1,10 Ketone bodies are generated in the liver and allow for the maintenance of systemic caloric and energy homeostasis during fasting and caloric restriction. It has previously been demonstrated that neonatal ketogenesis is activated independently of starvation. However, the role of ketogenesis during the perinatal period remains unclear. Here, we show that neonatal ketogen- esis plays a protective role in mitochondrial function. We generated a mouse model of insufficient ketogenesis by disrupting the rate-limiting hydroxymethylglutaryl-CoA synthase 2 enzyme gene (Hmgcs2). Hmgcs2 knockout (KO) neonates develop microvesicular steatosis within a few days of birth. Electron microscopic analysis and metabolite profiling indicate a restricted energy production capacity and accumulation of acetyl-CoA in Hmgcs2 KO mice. Furthermore, -
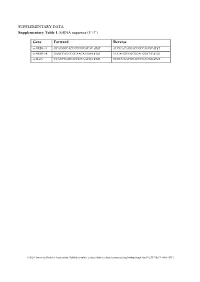
Supplementary Tables and Figures
SUPPLEMENTARY DATA Supplementary Table 1. SiRNA sequence (5’-3’) Gene Forward Reverse si-HRD1-1# GCAUGGCAGUCCUGUACAU dTdT AUGUACAGGACUGCCAUGC dTdT si-HRD1-2# GAGCCAUCCGCAACAUGAA dTdT UUCAUGUUGCGGAUGGCUC dTdT si-MafA CCAUCGAGUACGUCAACGA dTdT UCGUUGACGUACUCGAUGG dTdT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 2. Primer sequences for qRT-PCR (5’-3’) Gene Forward Reverse human HRD1 GCTCACGCCTACTACCTCAAA GCCAGACAAGTCTCTGTGACG mouse mafA AAGCGGCGCACGCTCAAGAA GGTCCCGCTCCTTGGCCAGA mouse insulin1 CACTTCCTACCCCTGCTGG ACCACAAAGATGCTGTTTGACA mouse β-actin AGGCCAACCGTGAAAAGATG AGAGCATAGCCCTCGTAGATGG human β-actin CATGTACGTTGCTATCCAGGC CTCCTTAATGTCACGCACGAT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 3. Primer sequences for ChIP (5’-3’) Gene promoter Forward Reverse mouse Insulin1, 2 GGAACTGTGAAACAGTCCAAGG CCCCCTGGACTTTGCTGTTTG ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 4. Primer sequences for PCR (5’-3’) Gene Forward Reverse HRD1-pDsred CCCAAGCTTATGTTCCGCACCGCAGT GGGGTACCCAGTGGGCAACAGGGG HRD1-pCMV- Flag GGGGTACCATGTTCCGCACCGCAGT CCCAAGCTTGTGGGCAACAGGGGACT C HRD1-pCMV-HA GGCCATGGGCCATATGGGATCCTTCC AGGGATGCCACCCGGGGATCCTCAGT GCACCGCAGTGATG GGGCAACAGGGGAC HRD1-N-HA GGCCATGGGCCATATGGGATCCTTCC -

Identification and Functional Characterization of the First Two
Identification and functional characterization of the first two aromatic prenyltransferases implicated in the biosynthesis of furanocoumarins and prenylated coumarins in two plant families: Rutaceae and Apiaceae Fazeelat Karamat To cite this version: Fazeelat Karamat. Identification and functional characterization of the first two aromatic prenyl- transferases implicated in the biosynthesis of furanocoumarins and prenylated coumarins in two plant families: Rutaceae and Apiaceae. Agronomy. Université de Lorraine, 2013. English. NNT : 2013LORR0029. tel-01749560 HAL Id: tel-01749560 https://hal.univ-lorraine.fr/tel-01749560 Submitted on 29 Mar 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. AVERTISSEMENT Ce document est le fruit d'un long travail approuvé par le jury de soutenance et mis à disposition de l'ensemble de la communauté universitaire élargie. Il est soumis à la propriété intellectuelle de l'auteur. Ceci implique une obligation de citation et de référencement lors de l’utilisation de ce document. D'autre part, toute contrefaçon, plagiat, -

KENNETH B. STOREY and ERNEST BAILEY Department of Biochemistry, University of Sheffield, Sheffield SI0 2TN, U.K
Insect Biochem.,. 1978, Vol. 8, pp. 125 to 131. Peroamon Press. Printed in Great Britain INTRACELLULAR DISTRIBUTION OF ENZYMES ASSOCIATED WITH LIPOGENESIS AND GLUCONEOGENESIS IN FAT BODY OF THE ADULT COCKROACH, PERIPLANETA KENNETH B. STOREY and ERNEST BAILEY Department of Biochemistry, University of Sheffield, Sheffield SI0 2TN, U.K. (Received 30 April 1977) Abstract--The intracellular distribution and maximal activities of some enzymes associated with lipo- genesis, gluconeogenesis and fatty acid oxidation have been determined in the fat body of the adult male cockroach Periplaneta americana. Of the enzymes of lipogenesis, acetyl-CoA synthase, acetyl-CoA carboxylase, glucose 6-phosphate dehydrogenase, and ATP citrate lyase are located entirely in the cytosol. Of the other enzymes of citrate metabolism studied, citrate synthase and NAD-dependent isocitrate dehydrogenase are almost exclusively mitochondrial, whereas NADP-dependent isocitrate dehydrogenase and aconitase are predominantly cytosolic although significant mitochondrial activity is also present. The latter subeellular distribution is also observed for 'malic enzyme' and NAD and NADP dependent malate dehydrogenase. The enzyme of fatty acid oxidation studied, 3-hydroxyacyl- CoA dehydrogenase is entirely mitochondrial. Of the enzymes possibly involved in glueoneogenesis, glucose 6-phosphate is microsomal, fructose 1,6-diphosphatase cytosolic, pyruvate carboxylase mito- chondrial and phosphoenolpyruvate carboxykinase predominantly eytosolic. Of the enzymes of amino acid metabolism studied, glutamate dehydrogenase is NAD-dependent and located in the mitochondria whereas glutamate/oxalacetate and glutamate/pyruvate transaminases are predominantly cytosolic but with significant activity in the mitochondria. Glycerol kinase and sorbitol dehydrogenase are cytosolic and the glyoxylate cycle enzymes malate synthetase and isocitrate lyase are not detected in the fat body. -

Candidate Genes Linked to QTL Regions Associated with Fatty Acid
1 Candidate genes linked to QTL regions associated with fatty acid composition in oil palm 2 3 Abstract 4 5 The present study searched for candidate genes in five linkage groups (LGs) - T2, T3, OT4, OT6 and T9 hosting the QTLs 6 associated with iodine value (IV) and fatty acid composition (FAC) in an oil palm interspecific hybrid population. Each 7 of the five LGs was successfully anchored to its corresponding chromosomal segment where, a wider repertoire of 8 candidate genes was identified. This study further revealed a total of 19 candidate genes and four transcription factors 9 involved in biosynthesis of fatty acids, lipids (including triacylglycerol) and acetyl-CoA, glycosylation and degradation 10 of fatty acids. Their possible involvement in regulating the levels of saturation are discussed. In addition, 22 candidate 11 genes located outside the QTL intervals were also identified across the interspecific hybrid genome. A total of 92 SSR 12 markers were developed to tag the presence of these candidate genes and 50 were successfully mapped onto their 13 respective positions on the genome. The data obtained here complements the previous studies, and collectively, these 14 QTL-linked candidate gene markers could help breeders in more precisely selecting palms with the desired FAC. 15 16 Keywords 17 18 Elaeis guineensis, interspecific hybrid, fatty acid biosynthesis, triacylglycerol biosynthesis, transcription factors, genetic 19 linkage map 20 21 Abbreviations 22 23 4CLL1 4-coumarate--CoA ligase 1 24 AACT acetoacetyl-CoA thiolase 25 acbd4 acyl-CoA-binding domain-containing protein 4 26 ACX4 acyl-CoA oxidase 4 27 C14:0 myristic acid 28 C16:0 palmitic acid 29 C16:1 palmitoleic acid 30 C18:0 stearic acid 31 C18:1 oleic acid 32 C18:2 linoleic acid 33 CHR pseudo-chromosome 34 cM centiMorgan 35 CP cross pollinator 36 CPO crude palm oil 37 CTAB cetyltriammonium bromide 38 DNA deoxyribonucleic acid 39 EG5 E.