Molecular Phylogeny of Ranunculaceae Based on Internal Transcribed Spacer Sequences
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Natural Communities of Michigan: Classification and Description
Natural Communities of Michigan: Classification and Description Prepared by: Michael A. Kost, Dennis A. Albert, Joshua G. Cohen, Bradford S. Slaughter, Rebecca K. Schillo, Christopher R. Weber, and Kim A. Chapman Michigan Natural Features Inventory P.O. Box 13036 Lansing, MI 48901-3036 For: Michigan Department of Natural Resources Wildlife Division and Forest, Mineral and Fire Management Division September 30, 2007 Report Number 2007-21 Version 1.2 Last Updated: July 9, 2010 Suggested Citation: Kost, M.A., D.A. Albert, J.G. Cohen, B.S. Slaughter, R.K. Schillo, C.R. Weber, and K.A. Chapman. 2007. Natural Communities of Michigan: Classification and Description. Michigan Natural Features Inventory, Report Number 2007-21, Lansing, MI. 314 pp. Copyright 2007 Michigan State University Board of Trustees. Michigan State University Extension programs and materials are open to all without regard to race, color, national origin, gender, religion, age, disability, political beliefs, sexual orientation, marital status or family status. Cover photos: Top left, Dry Sand Prairie at Indian Lake, Newaygo County (M. Kost); top right, Limestone Bedrock Lakeshore, Summer Island, Delta County (J. Cohen); lower left, Muskeg, Luce County (J. Cohen); and lower right, Mesic Northern Forest as a matrix natural community, Porcupine Mountains Wilderness State Park, Ontonagon County (M. Kost). Acknowledgements We thank the Michigan Department of Natural Resources Wildlife Division and Forest, Mineral, and Fire Management Division for funding this effort to classify and describe the natural communities of Michigan. This work relied heavily on data collected by many present and former Michigan Natural Features Inventory (MNFI) field scientists and collaborators, including members of the Michigan Natural Areas Council. -

Ranunculaceae) for Asian and North American Taxa
Mosyakin, S.L. 2018. Further new combinations in Anemonastrum (Ranunculaceae) for Asian and North American taxa. Phytoneuron 2018-55: 1–11. Published 13 August 2018. ISSN 2153 733X FURTHER NEW COMBINATIONS IN ANEMONASTRUM (RANUNCULACEAE) FOR ASIAN AND NORTH AMERICAN TAXA SERGEI L. MOSYAKIN M.G. Kholodny Institute of Botany National Academy of Sciences of Ukraine 2 Tereshchenkivska Street Kiev (Kyiv), 01004 Ukraine [email protected] ABSTRACT Following the proposed re-circumscription of genera in the group of Anemone L. and related taxa of Ranunculaceae (Mosyakin 2016, Christenhusz et al. 2018) and based on recent molecular phylogenetic and partly morphological evidence, the genus Anemonastrum Holub is recognized here in an expanded circumscription (including Anemonidium (Spach) Holub, Arsenjevia Starod., Tamuria Starod., and Jurtsevia Á. Löve & D. Löve) covering members of the “Anemone ” clade with x=7, but excluding Hepatica Mill., a genus well outlined morphologically and forming a separate subclade (accepted by Hoot et al. (2012) as Anemone subg. Anemonidium (Spach) Juz. sect. Hepatica (Mill.) Spreng.) within the clade earlier recognized taxonomically as Anemone subg. Anemonidium (sensu Hoot et al. 2012). The following new combinations at the section and subsection ranks are validated: Anemonastrum Holub sect. Keiskea (Tamura) Mosyakin, comb. nov . ( Anemone sect. Keiskea Tamura); Anemonastrum [sect. Keiskea ] subsect. Keiskea (Tamura) Mosyakin, comb. nov .; Anemonastrum [sect. Keiskea ] subsect. Arsenjevia (Starod.) Mosyakin, comb. nov . ( Arsenjevia Starod.); and Anemonastrum [sect. Anemonastrum ] subsect. Himalayicae (Ulbr.) Mosyakin, comb. nov. ( Anemone ser. Himalayicae Ulbr.). The new nomenclatural combination Anemonastrum deltoideum (Hook.) Mosyakin, comb. nov . ( Anemone deltoidea Hook.) is validated for a North American species related to East Asian Anemonastrum keiskeanum (T. -

Etude Sur L'origine Et L'évolution Des Variations Florales Chez Delphinium L. (Ranunculaceae) À Travers La Morphologie, L'anatomie Et La Tératologie
Etude sur l'origine et l'évolution des variations florales chez Delphinium L. (Ranunculaceae) à travers la morphologie, l'anatomie et la tératologie : 2019SACLS126 : NNT Thèse de doctorat de l'Université Paris-Saclay préparée à l'Université Paris-Sud ED n°567 : Sciences du végétal : du gène à l'écosystème (SDV) Spécialité de doctorat : Biologie Thèse présentée et soutenue à Paris, le 29/05/2019, par Felipe Espinosa Moreno Composition du Jury : Bernard Riera Chargé de Recherche, CNRS (MECADEV) Rapporteur Julien Bachelier Professeur, Freie Universität Berlin (DCPS) Rapporteur Catherine Damerval Directrice de Recherche, CNRS (Génétique Quantitative et Evolution Le Moulon) Présidente Dario De Franceschi Maître de Conférences, Muséum national d'Histoire naturelle (CR2P) Examinateur Sophie Nadot Professeure, Université Paris-Sud (ESE) Directrice de thèse Florian Jabbour Maître de conférences, Muséum national d'Histoire naturelle (ISYEB) Invité Etude sur l'origine et l'évolution des variations florales chez Delphinium L. (Ranunculaceae) à travers la morphologie, l'anatomie et la tératologie Remerciements Ce manuscrit présente le travail de doctorat que j'ai réalisé entre les années 2016 et 2019 au sein de l'Ecole doctorale Sciences du végétale: du gène à l'écosystème, à l'Université Paris-Saclay Paris-Sud et au Muséum national d'Histoire naturelle de Paris. Même si sa réalisation a impliqué un investissement personnel énorme, celui-ci a eu tout son sens uniquement et grâce à l'encadrement, le soutien et l'accompagnement de nombreuses personnes que je remercie de la façon la plus sincère. Je remercie très spécialement Florian Jabbour et Sophie Nadot, mes directeurs de thèse. -

Extended Phylogeny of Aquilegia: the Biogeographical and Ecological Patterns of Two Simultaneous but Contrasting Radiations
Plant Syst Evol (2010) 284:171–185 DOI 10.1007/s00606-009-0243-z ORIGINAL ARTICLE Extended phylogeny of Aquilegia: the biogeographical and ecological patterns of two simultaneous but contrasting radiations Jesu´s M. Bastida • Julio M. Alca´ntara • Pedro J. Rey • Pablo Vargas • Carlos M. Herrera Received: 29 April 2009 / Accepted: 25 October 2009 / Published online: 4 December 2009 Ó Springer-Verlag 2009 Abstract Studies of the North American columbines respective lineages. The genus originated between 6.18 (Aquilegia, Ranunculaceae) have supported the view that and 6.57 million years (Myr) ago, with the main pulses of adaptive radiations in animal-pollinated plants proceed diversification starting around 3 Myr ago both in Europe through pollinator specialisation and floral differentiation. (1.25–3.96 Myr ago) and North America (1.42–5.01 Myr However, although the diversity of pollinators and floral ago). The type of habitat occupied shifted more often in morphology is much lower in Europe and Asia than in the Euroasiatic lineage, while pollination vectors shifted North America, the number of columbine species is more often in the Asiatic-North American lineage. similar in the three continents. This supports the Moreover, while allopatric speciation predominated in the hypothesis that habitat and pollinator specialisation have European lineage, sympatric speciation acted in the North contributed differently to the radiation of columbines in American one. In conclusion, the radiation of columbines different continents. To establish the basic background to in Europe and North America involved similar rates of test this hypothesis, we expanded the molecular phylog- diversification and took place simultaneously and inde- eny of the genus to include a representative set of species pendently. -

Winter 2014-2015 (22:3) (PDF)
Contents NATIVE NOTES Page Fern workshop 1-2 Wavey-leaf basket Grass 3 Names Cacalia 4 Trip Report Sandstone Falls 5 Kate’s Mountain Clover* Trip Report Brush Creek Falls 6 Thank yous memorial 7 WEST VIRGINIA NATIVE PLANT SOCIETY NEWSLETTER News of WVNPS 8 VOLUME 22:3 WINTER 2014-15 Events, Dues Form 9 Judy Dumke-Editor: [email protected] Phone 740-894-6859 Magnoliales 10 e e e visit us at www.wvnps.org e e e . Fern Workshop University of Charleston Charleston WV January 17 2015, bad weather date January 24 2015 If you have thought about ferns, looked at them, puzzled over them or just want to know more about them join the WVNPS in Charleston for a workshop led by Mark Watson of the University of Charleston. The session will start at 10 A.M. with a scheduled end point by 12:30 P.M. A board meeting will follow. The sessions will be held in the Clay Tower Building (CTB) room 513, which is the botany lab. If you have any pressed specimens to share, or to ask about, be sure to bring them with as much information as you have on the location and habitat. Even photographs of ferns might be of interest for the session. If you have a hand lens that you favor bring it along as well. DIRECTIONS From the North: Travel I-77 South or 1-79 South into Charleston. Follow the signs to I-64 West. Take Oakwood Road Exit 58A and follow the signs to Route 61 South (MacCorkle Ave.). -
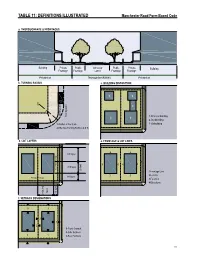
Manchester Road Redevelopment District: Form-Based Code
TaBle 11: deFiniTionS illuSTraTed manchester road Form-Based Code a. ThoroughFare & FronTageS Building Private Public Vehicular Public Private Building Frontage Frontage Lanes Frontage Frontage Private lot Thoroughfare (r.o.w.) Private lot b. Turning radiuS c. Building diSPoSiTion 3 3 2 2 1 Parking Lane Moving Lane 1- Principal Building 1 1 2- Backbuilding 1-Radius at the Curb 3- Outbuilding 2-Effective Turning Radius (± 8 ft) d. loT LAYERS e. FronTage & loT lineS 4 3rd layer 4 2 1 4 4 4 3 2nd layer Secondary Frontage 20 feet 1-Frontage Line 2-Lot Line 1st layer 3 3 Principal Frontage 3-Facades 1 1 4-Elevations layer 1st layer 2nd & 3rd & 2nd f. SeTBaCk deSignaTionS 3 3 2 1 2 1-Front Setback 2-Side Setback 1 1 3-Rear Setback 111 Manchester Road Form-Based Code ARTICLE 9. APPENDIX MATERIALS MBG Kemper Center PlantFinder About PlantFinder List of Gardens Visit Gardens Alphabetical List Common Names Search E-Mail Questions Menu Quick Links Home Page Your Plant Search Results Kemper Blog PlantFinder Please Note: The following plants all meet your search criteria. This list is not necessarily a list of recommended plants to grow, however. Please read about each PF Search Manchesterplant. Some may Road be invasive Form-Based in your area or may Code have undesirable characteristics such as above averageTab insect LEor disease 11: problems. NATIVE PLANT LIST Pests Plants of Merit Missouri Native Plant List provided by the Missouri Botanical Garden PlantFinder http://www.mobot.org/gardeninghelp/plantfinder Master Search Search limited to: Missouri Natives Search Tips Scientific Name Scientific Name Common NameCommon Name Height (ft.) ZoneZone GardeningHelp (ft.) Acer negundo box elder 30-50 2-10 Acer rubrum red maple 40-70 3-9 Acer saccharinum silver maple 50-80 3-9 Titles Acer saccharum sugar maple 40-80 3-8 Acer saccharum subsp. -

Ranunculaceae – Buttercup Family
RANUNCULACEAE – BUTTERCUP FAMILY Plant: mostly herbs, some woody vines or shrubs Stem: Root: Leaves: mostly alternate, sometimes opposite or whorled or basal; lobed or not lobed; if lobed then most often palmately, but occasionally pinnately, sometimes finely dissected – highly variable, sometimes even on the same plant; with or without stipules Flowers: mostly perfect, some dioecious; sepals 3-6, commonly 5; petals vary in number (3-23) but often 5, petals may be lacking and sepals are showy; stamens few to many; ovary superior, carpels few to very many, pistils one to many Fruit: mostly a dry capsule, seeds small, may be oily; rarely a berry Other: large family, sometimes confused with members of the Rose family (5 petals); Dicotyledons Group Genera: 60+ genera; locally Actaea (baneberry), Anemone (anemone or windflower), Aquilegia (columbine), Clematis, Isopyrum, Hepatica, Hydrastis, Ranunuculus (buttercup or crowfoot), Thalictrum (meadow-rue) WARNING – family descriptions are only a layman’s guide and should not be used as definitive Flower Morphology in the This is a large family often based on 5’s but Ranunculaceae (Buttercup Family) exceptions occur Examples of common genera White Baneberry [Doll’s-Eyes] Yellow Marsh Marigold [Cowslip] Goldenseal [Yellowroot] Actaea pachypoda Ell. Carolina [Wild Blue] Larkspur Caltha palustris L. var. palustris Delphinium carolinianum Walter Hydrastis canadensis L. Swamp Leather Flower [Eastern] False Rue Anemone Clematis crispa L. Devil-In-The-Bush [Love American Wood Anemone Enemion biternatum Raf. -In-A-Mist] Anemone quinquefolia L. [Isopyrum biternatum] Nigella damascena L. (Introduced) Doubtful [Rocket; Garden] Knight's-Spur [Larkspur] Round-lobed Hepatica [Liverleaf] Tall Buttercup Hepatica nobilis Schreber var. -

Plant Introductions
Plant Introductions Promoting the garden use of native plants is a core part of Mt. Cuba Center’s mission, and since 1988, the Plant Introduction Program has brought many outstanding cultivars to market. Selected for strong ornamental appeal and broad regional adaptability, many of these introductions have risen to become some of the most popular cultivars of native plants available. Plant evaluation and introduction continues today, with several bright prospects for additional introductions in the future. Plant Introductions Actaea pachypoda ‘Misty Blue’ Misty Blue white baneberry Misty Blue white baneberry is an herbaceous perennial, selected for its unique and highly attractive soft, bluish-green foliage. It was discovered in a planting of typical green-leaved plants of unknown origin growing at Mt. Cuba Center. In spring 1" to 2" tall bottlebrush-like clusters of white flowers are borne on stems above the foliage. By September, large, white fruit with dark purple to black spots (“doll’s eyes”) mature on reddish pedicels. It is a carefree, long-lived, 24" to 36" tall plant, growing best in partial to filtered shade in evenly moist, well-drained soils with a pH from slightly acidic to neutral. Introduced 2009 Ageratina altissima ‘Chocolate’ (formerly Eupatorium rugosum) Chocolate white snakeroot Chocolate white snakeroot is an herbaceous perennial selected by Dr. Richard W. Lighty and descended from a plant found at Winterthur Gardens. It had the darkest burgundy foliage of many seedlings grown over a ten year period. ‘Chocolate’ grows up to 3' tall and has white inflorescences along with dark burgundy leaves which color best in full sun. -

And Natural Community Restoration
RECOMMENDATIONS FOR LANDSCAPING AND NATURAL COMMUNITY RESTORATION Natural Heritage Conservation Program Wisconsin Department of Natural Resources P.O. Box 7921, Madison, WI 53707 August 2016, PUB-NH-936 Visit us online at dnr.wi.gov search “ER” Table of Contents Title ..……………………………………………………….……......………..… 1 Southern Forests on Dry Soils ...................................................... 22 - 24 Table of Contents ...……………………………………….….....………...….. 2 Core Species .............................................................................. 22 Background and How to Use the Plant Lists ………….……..………….….. 3 Satellite Species ......................................................................... 23 Plant List and Natural Community Descriptions .…………...…………….... 4 Shrub and Additional Satellite Species ....................................... 24 Glossary ..................................................................................................... 5 Tree Species ............................................................................... 24 Key to Symbols, Soil Texture and Moisture Figures .................................. 6 Northern Forests on Rich Soils ..................................................... 25 - 27 Prairies on Rich Soils ………………………………….…..….……....... 7 - 9 Core Species .............................................................................. 25 Core Species ...……………………………….…..…….………........ 7 Satellite Species ......................................................................... 26 Satellite Species -

2021 Plant List
2021 Plant List New items are listed with an asterisk (*) Conifers Pinus thungerbii Abies koreana 'Horstmann's Silberlocke' Pinus x 'Jane Kluis' * Chamaecyparis nootkatensis 'Pendula' Sciadopitys vert. 'Joe Dozey' Chamaecyparis noot. 'Glauca Pendula' Sciadopitys vert. 'Wintergreen' Chamaecyparis obtusa 'Chirimen' * Taxodium distichum 'Pendula' Chamaecyparis obtusa 'Gracilis' -Select Taxodium distichum 'Peve Mineret' Chamaecyparis obtusa 'Kosteri' Taxus cuspidaata 'Nana Aurescens' Chamaecyparis obtusa 'Nana' Tsuga con. 'Jervis' Chamaecyparis obtusa 'Nana Gracilis' Chamaecyparis obtusa 'Spiralis' Ferns Chamaecyparis obtusa 'Thoweil' Adiantum pedatum ….Maiden Hair Chamaecyparis obtusa 'Verdoni' Athyrum filix-femina 'Minutissima' Juniperus procumbens 'Nana' Athyrium 'Ghost' Larix decidua 'Pendula' Athyrum niponicum 'Godzilla' Larix decidua 'Pendula' -Prostrate Form Athyrum niponicum 'Pictum' Picea abies 'Hasin' * Athyrum niponicum pic. 'Pearly White' Picea abies 'Pusch' * Dennstaedtia punctilobula Picea omorika 'Nana' Dryopteris ery. 'Brilliance' Picea omorika 'Pendula' Dryopteris marginalis Picea orientalis 'Nana' Matteucciastruthiopteris var. pensylvanica Picea orientalis 'Shadow's Broom' * Osmunda cinnamomea Picea pungens 'Glauca Globosa' Polystichum acrostichoides Pinus mugo 'Mughus' - Rock Garden Strain Polystichum polyblepharum Pinus mugo 'Slowmound' Pinus nigra 'Hornibrookiana' Grasses Pinus parviflora 'Aoi' These are but a fraction of the grasses we'll be Pinus parviflora 'Glauca Nana' offering this year. Many more to come. They'll -

List of Plants for Great Sand Dunes National Park and Preserve
Great Sand Dunes National Park and Preserve Plant Checklist DRAFT as of 29 November 2005 FERNS AND FERN ALLIES Equisetaceae (Horsetail Family) Vascular Plant Equisetales Equisetaceae Equisetum arvense Present in Park Rare Native Field horsetail Vascular Plant Equisetales Equisetaceae Equisetum laevigatum Present in Park Unknown Native Scouring-rush Polypodiaceae (Fern Family) Vascular Plant Polypodiales Dryopteridaceae Cystopteris fragilis Present in Park Uncommon Native Brittle bladderfern Vascular Plant Polypodiales Dryopteridaceae Woodsia oregana Present in Park Uncommon Native Oregon woodsia Pteridaceae (Maidenhair Fern Family) Vascular Plant Polypodiales Pteridaceae Argyrochosma fendleri Present in Park Unknown Native Zigzag fern Vascular Plant Polypodiales Pteridaceae Cheilanthes feei Present in Park Uncommon Native Slender lip fern Vascular Plant Polypodiales Pteridaceae Cryptogramma acrostichoides Present in Park Unknown Native American rockbrake Selaginellaceae (Spikemoss Family) Vascular Plant Selaginellales Selaginellaceae Selaginella densa Present in Park Rare Native Lesser spikemoss Vascular Plant Selaginellales Selaginellaceae Selaginella weatherbiana Present in Park Unknown Native Weatherby's clubmoss CONIFERS Cupressaceae (Cypress family) Vascular Plant Pinales Cupressaceae Juniperus scopulorum Present in Park Unknown Native Rocky Mountain juniper Pinaceae (Pine Family) Vascular Plant Pinales Pinaceae Abies concolor var. concolor Present in Park Rare Native White fir Vascular Plant Pinales Pinaceae Abies lasiocarpa Present -

Black Cohosh & Endangered Species Actaea Racemosa L
Natural Heritage Black Cohosh & Endangered Species Actaea racemosa L. Program State Status: Endangered www.mass.gov/nhesp Federal Status: None Massachusetts Division of Fisheries & Wildlife DESCRIPTION: Black Cohosh (Actaea racemosa, formerly Cimicifuga racemosa) is a striking herbaceous perennial plant of the buttercup family (Ranunculaceae), with alternate, compound leaves and four to nine malodorous, wand-like, white inflorescences. Though indigenous to rich woodlands, Black Cohosh is also a common garden and herbal medicinal plant, and goes by the other common names Black Snakeroot, Squawroot, and Bugbane. AIDS TO IDENTIFICATION: The leaves of Black Cohosh are 15 to 60 cm (~6–23 in.) in length, smooth, and two to three times “ternately” (i.e., divided in three) compound, with 20 to 70 toothed leaflets. The flowering stem can be quite tall, reaching up to 2.5 m (~8 ft.) in height; it is branched, with several racemes of fetid, white flowers. Individual flowers appear as a mass of stamens with white filaments 5 to 10 mm long, topped by rounded anthers. The fruit is a thick-walled follicle, 5 to 10 mm in size. SIMILAR SPECIES: The leaves of Black Cohosh resemble those of Red Baneberry (Actaea rubra), and White Baneberry (Actaea pachypoda). Like Black Cohosh, baneberries are known from rich woodlands and have compound leaves with toothed leaflets, but they are typically much smaller plants. The most distinguishing characters are the inflorescence and the fruit; in baneberries, the inflorescence is an unbranched raceme, and the fruit are berries, not follicles. HABITAT: In Massachusetts, Black Cohosh inhabits very rich deciduous forests typically with moist alkaline soils.