Seasonal Monitoring of Hida Salamander Hynobius Kimurae Using Environmental DNA with a Genus-Specific Primer Set
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A New Cascade Frog of the Subgenus Odorrana from Peninsular Malaysia
ZOOLOGICAL SCIENCE 23: 647–651 (2006) 2006 Zoological Society of Japan A New Cascade Frog of the Subgenus Odorrana from Peninsular Malaysia Masafumi Matsui1* and Ibrahim Jaafar 2 1Graduate School of Human and Environmental Studies, Kyoto University, Sakyo-ku, Kyoto 606-8501, Japan 2Biological Sciences Program, School of Distance Education, Universiti Sains Malaysia, 11800 USM, Penang, Malaysia We describe a new species of cascade frog of the genus Rana, from west Malaysia. Rana monjerai, new species is a medium-sized frog of the subgenus Odorrana (SVL of males, 38–43 mm; of one female, 75 mm), and is distinguished from all other members of this subgenus by the combination of: white lip stripe, dorsolateral fold, full web on the fourth toe, vomerine teeth, gular vocal pouch and relatively large tympanum in males, no dorsal marking, no clear light spots on rear of thigh, first finger subequal to second, finely tuberculated dorsum, and unpigmented ova. The significance of finding this species from peninsular Malaysia is discussed. Key words: cryptic species, Rana, new species, Southeast Asia, taxonomy, zoogeography ficially resembling R. hosii, which the senior author (Matsui, INTRODUCTION unpublished data) had obtained at higher elevations on the Along mountain streams in subtropical and tropical same mountain. Later examination of these specimens, regions of East to Southeast Asia, there are small to however, revealed that they are clearly different from R. medium-sized, long-legged ranid frogs represented by spe- hosii in the presence of gular pouches in males. Further cies like R. narina Stejneger, 1901 from the Ryukyu Archi- study of the specimens by consulting with a recent review of pelago of Japan; R. -

Caudata: Hynobiidae): Heterochronies and Reductions
65 (1): 117 – 130 © Senckenberg Gesellschaft für Naturforschung, 2015. 4.5.2015 Development of the bony skeleton in the Taiwan salamander, Hynobius formosanus Maki, 1922 (Caudata: Hynobiidae): Heterochronies and reductions Anna B. Vassilieva 1 *, June-Shiang Lai 2, Shang-Fang Yang 2, Yu-Hao Chang 1 & Nikolay A. Poyarkov, Jr. 1 1 Department of Vertebrate Zoology, Biological Faculty, Lomonosov Moscow State University, Leninskiye Gory, GSP-1, Moscow 119991, Russia — 2 Department of Life Science, National Taiwan Normal University, 88, Sec. 4 Tingchou Rd., Taipei 11677, Taiwan, R.O.C. — *Cor- responding author; vassil.anna(at)gmail.com Accepted 19.ii.2015. Published online at www.senckenberg.de / vertebrate-zoology on 4.v.2015. Abstract The development of the bony skeleton in a partially embryonized lotic-breeding salamander Hynobius formosanus is studied using the ontogenetic series from late embryos to postmetamorphic juveniles and adult specimen. Early stages of skull development in this spe- cies are compared with the early cranial ontogeny in two non-embryonized lentic-breeding species H. lichenatus and H. nigrescens. The obtained results show that skeletal development distinguishes H. formosanus from other hynobiids by a set of important features: 1) the reduction of provisory ossifications (complete absence of palatine and reduced state of coronoid), 2) alteration of a typical sequence of ossification appearance, namely, the delayed formation of vomer and coronoid, and 3) the absence of a separate ossification center of a lacrimal and formation of a single prefrontolacrimal. These unique osteological characters in H. formosanus are admittedly connected with specific traits of its life history, including partial embryonization, endogenous feeding until the end of metamorphosis and relatively short larval period. -

Zootaxa,Paraphyly of Chinese Amolops (Anura, Ranidae) and Phylogenetic Position of The
Zootaxa 1531: 49–55 (2007) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ ZOOTAXA Copyright © 2007 · Magnolia Press ISSN 1175-5334 (online edition) Paraphyly of Chinese Amolops (Anura, Ranidae) and phylogenetic position of the rare Chinese frog, Amolops tormotus HONG-XIA CAI1, 2, JING CHE2, JUN-FENG PANG2, ER-MI ZHAO1,4& YA-PING ZHANG2, 3,4 1Key Laboratory of Bio-resources and Eco-environment (Ministry of Education), College of Life Sciences, Sichuan University, Chengdu, China, 610064 2Laboratory of Cellular and Molecular Evolution, Kunming Institute of Zoology, the Chinese Academy of Sciences, Kunming, China, 650223 3Laboratory for Conservation and Utilization of Bio-resources, Yunnan University, Kunming, China, 650091 4Corresponding authors. E-mail: [email protected]; [email protected] Abstract In order to evaluate the five species groups of Chinese Amolops based on morphological characteristics, and to clarify the phylogenetic position of the concave-eared torrent frog Amolops tormotus, we investigated the phylogeny of Amolops by maximum parsimony, Bayesian Inference, and maximum likelihood methods using two mitochondrial DNA fragments (12S rRNA, 16S rRNA). Our results supported a sister group relationship of Amolops ricketti and Amolops hainanensis. However, the grouping of Amolops mantzorum and Amolops monticola needs to be resolved with more data. Amolops tormotus was nested in genus Odorrana. Thus, recognition of the A. tormotus group is unwarranted and A. tormotus should be referred to genus Odorrana as O. tormota. This species is the sister group of O. nasica plus O. versabilis. The new classification implies that the genus Wurana is to be considered as junior subjective synonym of Odorrana. -

About the Book the Format Acknowledgments
About the Book For more than ten years I have been working on a book on bryophyte ecology and was joined by Heinjo During, who has been very helpful in critiquing multiple versions of the chapters. But as the book progressed, the field of bryophyte ecology progressed faster. No chapter ever seemed to stay finished, hence the decision to publish online. Furthermore, rather than being a textbook, it is evolving into an encyclopedia that would be at least three volumes. Having reached the age when I could retire whenever I wanted to, I no longer needed be so concerned with the publish or perish paradigm. In keeping with the sharing nature of bryologists, and the need to educate the non-bryologists about the nature and role of bryophytes in the ecosystem, it seemed my personal goals could best be accomplished by publishing online. This has several advantages for me. I can choose the format I want, I can include lots of color images, and I can post chapters or parts of chapters as I complete them and update later if I find it important. Throughout the book I have posed questions. I have even attempt to offer hypotheses for many of these. It is my hope that these questions and hypotheses will inspire students of all ages to attempt to answer these. Some are simple and could even be done by elementary school children. Others are suitable for undergraduate projects. And some will take lifelong work or a large team of researchers around the world. Have fun with them! The Format The decision to publish Bryophyte Ecology as an ebook occurred after I had a publisher, and I am sure I have not thought of all the complexities of publishing as I complete things, rather than in the order of the planned organization. -

The Spemann Organizer Meets the Anterior‐
The Japanese Society of Developmental Biologists Develop. Growth Differ. (2015) 57, 218–231 doi: 10.1111/dgd.12200 Original Article The Spemann organizer meets the anterior-most neuroectoderm at the equator of early gastrulae in amphibian species Takanori Yanagi,1,2† Kenta Ito,1,2† Akiha Nishihara,1† Reika Minamino,1,2 Shoko Mori,1 Masayuki Sumida3 and Chikara Hashimoto1,2* 1JT Biohistory Research Hall, 1-1 Murasaki-cho, Takatsuki, Osaka 569-1125, 2Department of Biological Sciences, Graduate School of Science, Osaka University, Toyonaka, Osaka 560-0043, and 3Institute for Amphibian Biology, Hiroshima University, Kagamiyama, Higashi-Hiroshima, Hiroshima 739-8526, Japan The dorsal blastopore lip (known as the Spemann organizer) is important for making the body plan in amphibian gastrulation. The organizer is believed to involute inward and migrate animally to make physical contact with the prospective head neuroectoderm at the blastocoel roof of mid- to late-gastrula. However, we found that this physical contact was already established at the equatorial region of very early gastrula in a wide variety of amphibian species. Here we propose a unified model of amphibian gastrulation movement. In the model, the organizer is present at the blastocoel roof of blastulae, moves vegetally to locate at the region that lies from the blastocoel floor to the dorsal lip at the onset of gastrulation. The organizer located at the blastocoel floor con- tributes to the anterior axial mesoderm including the prechordal plate, and the organizer at the dorsal lip ends up as the posterior axial mesoderm. During the early step of gastrulation, the anterior organizer moves to estab- lish the physical contact with the prospective neuroectoderm through the “subduction and zippering” move- ments. -

50 CFR Ch. I (10–1–20 Edition) § 16.14
§ 15.41 50 CFR Ch. I (10–1–20 Edition) Species Common name Serinus canaria ............................................................. Common Canary. 1 Note: Permits are still required for this species under part 17 of this chapter. (b) Non-captive-bred species. The list 16.14 Importation of live or dead amphib- in this paragraph includes species of ians or their eggs. non-captive-bred exotic birds and coun- 16.15 Importation of live reptiles or their tries for which importation into the eggs. United States is not prohibited by sec- Subpart C—Permits tion 15.11. The species are grouped tax- onomically by order, and may only be 16.22 Injurious wildlife permits. imported from the approved country, except as provided under a permit Subpart D—Additional Exemptions issued pursuant to subpart C of this 16.32 Importation by Federal agencies. part. 16.33 Importation of natural-history speci- [59 FR 62262, Dec. 2, 1994, as amended at 61 mens. FR 2093, Jan. 24, 1996; 82 FR 16540, Apr. 5, AUTHORITY: 18 U.S.C. 42. 2017] SOURCE: 39 FR 1169, Jan. 4, 1974, unless oth- erwise noted. Subpart E—Qualifying Facilities Breeding Exotic Birds in Captivity Subpart A—Introduction § 15.41 Criteria for including facilities as qualifying for imports. [Re- § 16.1 Purpose of regulations. served] The regulations contained in this part implement the Lacey Act (18 § 15.42 List of foreign qualifying breed- U.S.C. 42). ing facilities. [Reserved] § 16.2 Scope of regulations. Subpart F—List of Prohibited Spe- The provisions of this part are in ad- cies Not Listed in the Appen- dition to, and are not in lieu of, other dices to the Convention regulations of this subchapter B which may require a permit or prescribe addi- § 15.51 Criteria for including species tional restrictions or conditions for the and countries in the prohibited list. -
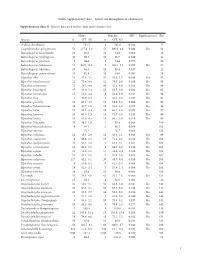
I Online Supplementary Data – Sexual Size Dimorphism in Salamanders
Online Supplementary data – Sexual size dimorphism in salamanders Supplementary data S1. Species data used in this study and references list. Males Females SSD Significant test Ref Species n SVL±SD n SVL±SD Andrias davidianus 2 532.5 8 383.0 -0.280 12 Cryptobranchus alleganiensis 53 277.4±5.2 52 300.9±3.4 0.084 Yes 61 Batrachuperus karlschmidti 10 80.0 10 84.8 0.060 26 Batrachuperus londongensis 20 98.6 10 96.7 -0.019 12 Batrachuperus pinchonii 5 69.6 5 74.6 0.070 26 Batrachuperus taibaiensis 11 92.9±12.1 9 102.1±7.1 0.099 Yes 27 Batrachuperus tibetanus 10 94.5 10 92.8 -0.017 12 Batrachuperus yenyuadensis 10 82.8 10 74.8 -0.096 26 Hynobius abei 24 57.8±2.1 34 55.0±1.2 -0.048 Yes 92 Hynobius amakusaensis 22 75.4±4.8 12 76.5±3.6 0.014 No 93 Hynobius arisanensis 72 54.3±4.8 40 55.2±4.8 0.016 No 94 Hynobius boulengeri 37 83.0±5.4 15 91.5±3.8 0.102 Yes 95 Hynobius formosanus 15 53.0±4.4 8 52.4±3.9 -0.011 No 94 Hynobius fuca 4 50.9±2.8 3 52.8±2.0 0.037 No 94 Hynobius glacialis 12 63.1±4.7 11 58.9±5.2 -0.066 No 94 Hynobius hidamontanus 39 47.7±1.0 15 51.3±1.2 0.075 Yes 96 Hynobius katoi 12 58.4±3.3 10 62.7±1.6 0.073 Yes 97 Hynobius kimurae 20 63.0±1.5 15 72.7±2.0 0.153 Yes 98 Hynobius leechii 70 61.6±4.5 18 66.5±5.9 0.079 Yes 99 Hynobius lichenatus 37 58.5±1.9 2 53.8 -0.080 100 Hynobius maoershanensis 4 86.1 2 80.1 -0.069 101 Hynobius naevius 72.1 76.7 0.063 102 Hynobius nebulosus 14 48.3±2.9 12 50.4±2.1 0.043 Yes 96 Hynobius osumiensis 9 68.4±3.1 15 70.2±3.0 0.026 No 103 Hynobius quelpaertensis 41 52.5±3.8 4 61.3±4.1 0.167 Yes 104 Hynobius -

Cfreptiles & Amphibians
WWW.IRCF.ORG TABLE OF CONTENTS IRCF REPTILES &IRCF AMPHIBIANS REPTILES • VOL &15, AMPHIBIANS NO 4 • DEC 2008 • 189 27(2):154–160 • AUG 2020 IRCF REPTILES & AMPHIBIANS CONSERVATION AND NATURAL HISTORY TABLE OF CONTENTS FEATURE ARTICLES A Herpetofaunal. Chasing Bullsnakes (Pituophis catenifer sayi) in Wisconsin: Survey of Northwestern On the Road to Understanding the Ecology and Conservation of the Midwest’s Giant Serpent ...................... Joshua M. Kapfer 190 Mongolia. The Shared History ofwith Treeboas (Corallus the grenadensis) andFirst Humans on Grenada: Country Record of A Hypothetical Excursion ............................................................................................................................Robert W. Henderson 198 theRESEARCH Moorfrog, ARTICLES Rana arvalis Nilsson 1842 . The Texas Horned Lizard in Central and Western Texas ....................... Emily Henry, Jason Brewer, Krista Mougey, and Gad Perry 204 Munkhbaatar .MunkhbayarThe Knight Anole1 (,Anolis Terbish equestris Khayankhyarvaa) in Florida 2, Onolragchaa Ganbold1, Zoljargal Purevdorj3, Burnee Mundur4, .............................................GurragchaaBrian J. Camposano, Jargalsaikhan Kenneth L. Krysko,1, and Kevin Munkhbayar M. Enge, Ellen M. Khorloo Donlan, and1 Michael Granatosky 212 1 DepartmentCONSERVATION of Biology, Mongolian ALERT National University of Education, Ulaanbaatar, Mongolia ([email protected]) . World’s2 DepartmentMammals in Crisis of Biology, .............................................................................................................................. -

Salamandrella Keyserlingii, Amphibia, Caudata) and the Cryptic Species S
Entomological Review, Vol. 85, Suppl. 2, 2005, pp. S240–S253. Translated from Zoologicheskii Zhurnal, Vol. 84, no. 11, 2005. Original Russian Text Copyright © 2005 by Berman, Derenko, Malyarchuk, Grzybowski, Kryukov, Miscicka-Sliwka. English Translation Copyright © 2005 by Pleiades Publishing, Inc. Intraspecific Genetic Differentiation of the Siberian Newt (Salamandrella keyserlingii, Amphibia, Caudata) and the Cryptic Species S. schrenckii from Southeastern Russia D. I. Berman*, M. V. Derenko*, B. A. Malyarchuk*, T. Grzybowski**, A. P. Kryukov***, and D. Miscicka-Sliwka** *Institute of Biological Problems of the North, Far East Division, Russian Academy of Sciences, Magadan, 685000 Russia e-mail: [email protected], [email protected] **Forensic Medicine Institute, Ludwik Rydygier Medical University, Bydgoszcz, 85-094 Poland ***Institute of Biology and Soil Science, Far East Division, Russian Academy of Sciences, Vladivostok, 690022 Russia Received March 16, 2005 Abstract—The nucleotide sequences of the mitochondrial cytochrome b gene in the Siberian newt Salaman- drella keyserlingii Dybowski 1870 from the populations of the Ural Mountains, Magadan oblast, Chukchi Pen- insula, Sakhalin Island, and Primorskii krai are analyzed. It is shown that in most populations studied (except for Primorskii krai), a low geographic variation in morphological characters corresponds to a low level of genetic variation (0.38% in the combined sample from the Magadan, Sakhalin, Chukchi, and Ural populations). Different scenarios for the origin of the genetically and morphologically homogeneous hyperpopulation are dis- cussed, taking into account the obvious lack of genetic exchange between the marginal populations of the range. They involve the rapid formation of the species range in the Holocene, which followed its gradual development in the Pleistocene; unidirectional stabilizing selection within the entire range; the maintenance of variation at a stable level by mixing of the population during the dispersal of the young and, possibly, by group fertilization. -

01 Hadijah Norhaslind.Pmd
Malays. Appl.AMPHIBIAN Biol. (2011) ASEMBLAGE 40(1): 1-6 OF BUBU PERMANENT FOREST RESERVE, PERAK, PENINSULAR MALAYSIA 1 AMPHIBIAN ASEMBLAGE OF BUBU PERMANENT FOREST RESERVE, PERAK, PENINSULAR MALAYSIA IBRAHIM, J.*, WONG, J., MOHD FAZLIN, M.S., FATAN, H.Y., SITI HADIJAH, Y. and NORHASLINDA, S. Biological Sciences Program, School of Distance Education, Universiti Sains Malaysia, 11800, Penang, Malaysia *E-mail: [email protected] ABSTRACT A brief study of the amphibian fauna of Bubu Permanent Forest Reserve, Perak, which constitutes the southernmost part of the Bintang Hijau Forest Reserve, was carried out from 18 to 21 December 2006 to determine the diversity and density estimate of amphibian species in the area. Field parties comprising seven persons searched and collected amphibians from a small stream, the Dal River, in Compartments 6 and 7 of the Bubu Permanent Forest Reserve, from 2000 hrs to 2300 hrs for three consecutive nights. A total of 79 individual frogs from 13 species in five families were recorded. The five most abundant species were Hyalarana nicobariensis, H. labialis, Pedostibes hosii, Phrynoides aspera and Amolops larutensis. Six other species were considered rare, namely, Limnonectes doriae, H. picturata, Odorrana hosii, Polypedates leucomystax and Rhacophorus bimaculatus. Preliminary analysis of the data showed that the estimated populations of H. nicobariensis, H. labialis and P. hosii are 23, 13 and 10 individuals per 100 meters of river length, respectively. Due to the brief nature of the study, and the small area covered, the number of species is considered relatively moderate. More than 95% of frogs collected are forest species that require clean and pollution-free habitats to survive. -

A New Species of Odorrana Inhabiting Complete Darkness in a Karst Cave in Guangxi, China
Asian Herpetological Research 2015, 6(1): 11–17 ORIGINAL ARTICLE DOI: 10.16373/j.cnki.ahr.140054 A New Species of Odorrana Inhabiting Complete Darkness in a Karst Cave in Guangxi, China Yunming MO1, Weicai CHEN1*, Huaying WU1, Wei ZHANG2 and Shichu ZHOU1 1 Natural History Museum of Guangxi, Nanning 530012, Guangxi, China 2 School of Life Sciences, East China Normal University, Shanghai 200062, China Abstract A new species of the genus Odorrana is described from a completely dark karst cave of northeastern Guangxi, southern China. The new species, Odorrana lipuensis sp. nov., can be distinguished from its congeners by a combination of the following characters: medium size (SVL: 40.7–47.7 mm in males, 51.1–55.4 mm in females); tips of all but first finger expanded with circummarginal grooves; smooth, grass-green dorsum with irregular brown mottling; pineal body invisible; throat to upper abdomen with gray mottling; dorsal surfaces of limbs with brown bands; dorsolateral fold absent; tiny spinules on lateral body, temporal region, and anterior and posterior edge of tympanum; white nuptial pad present on finger I; males lacking vocal sacs; females having creamy yellow eggs, without black poles. Uncorrected sequence divergences between O. lipuensis sp. nov. and all homologous 16S rRNA sequences of Odorrana available on GenBank is equal to or greater than 4.9%. Currently, the new species is only known from the type locality. Keywords Odorrana lipuensis sp. nov., karst cave, Guangxi, southern China 1. Introduction monophyletic group (Chen et al., 2013). All are known to be associated with mountain streams except O. -

Salamander Species Listed As Injurious Wildlife Under 50 CFR 16.14 Due to Risk of Salamander Chytrid Fungus Effective January 28, 2016
Salamander Species Listed as Injurious Wildlife Under 50 CFR 16.14 Due to Risk of Salamander Chytrid Fungus Effective January 28, 2016 Effective January 28, 2016, both importation into the United States and interstate transportation between States, the District of Columbia, the Commonwealth of Puerto Rico, or any territory or possession of the United States of any live or dead specimen, including parts, of these 20 genera of salamanders are prohibited, except by permit for zoological, educational, medical, or scientific purposes (in accordance with permit conditions) or by Federal agencies without a permit solely for their own use. This action is necessary to protect the interests of wildlife and wildlife resources from the introduction, establishment, and spread of the chytrid fungus Batrachochytrium salamandrivorans into ecosystems of the United States. The listing includes all species in these 20 genera: Chioglossa, Cynops, Euproctus, Hydromantes, Hynobius, Ichthyosaura, Lissotriton, Neurergus, Notophthalmus, Onychodactylus, Paramesotriton, Plethodon, Pleurodeles, Salamandra, Salamandrella, Salamandrina, Siren, Taricha, Triturus, and Tylototriton The species are: (1) Chioglossa lusitanica (golden striped salamander). (2) Cynops chenggongensis (Chenggong fire-bellied newt). (3) Cynops cyanurus (blue-tailed fire-bellied newt). (4) Cynops ensicauda (sword-tailed newt). (5) Cynops fudingensis (Fuding fire-bellied newt). (6) Cynops glaucus (bluish grey newt, Huilan Rongyuan). (7) Cynops orientalis (Oriental fire belly newt, Oriental fire-bellied newt). (8) Cynops orphicus (no common name). (9) Cynops pyrrhogaster (Japanese newt, Japanese fire-bellied newt). (10) Cynops wolterstorffi (Kunming Lake newt). (11) Euproctus montanus (Corsican brook salamander). (12) Euproctus platycephalus (Sardinian brook salamander). (13) Hydromantes ambrosii (Ambrosi salamander). (14) Hydromantes brunus (limestone salamander). (15) Hydromantes flavus (Mount Albo cave salamander).