Transforming Growth Factor-Β1 Promotes Breast Cancer Metastasis by Downregulating Mir-196A-3P Expression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Produzent Adresse Land Allplast Bangladesh Ltd
Zeitraum - Produzenten mit einem Liefertermin zwischen 01.01.2020 und 31.12.2020 Produzent Adresse Land Allplast Bangladesh Ltd. Mulgaon, Kaliganj, Gazipur, Rfl Industrial Park Rip, Mulgaon, Sandanpara, Kaligonj, Gazipur, Dhaka Bangladesh Bengal Plastics Ltd. (Unit - 3) Yearpur, Zirabo Bazar, Savar, Dhaka Bangladesh Durable Plastic Ltd. Mulgaon, Kaligonj, Gazipur, Dhaka Bangladesh HKD International (Cepz) Ltd. Plot # 49-52, Sector # 8, Cepz, Chittagong Bangladesh Lhotse (Bd) Ltd. Plot No. 60 & 61, Sector -3, Karnaphuli Export Processing Zone, North Potenga, Chittagong Bangladesh Plastoflex Doo Branilaca Grada Bb, Gračanica, Federacija Bosne I H Bosnia-Herz. ASF Sporting Goods Co., Ltd. Km 38.5, National Road No. 3, Thlork Village, Chonrok Commune, Konrrg Pisey, Kampong Spueu Cambodia Powerjet Home Product (Cambodia) Co., Ltd. Manhattan (Svay Rieng) Special Economic Zone, National Road 1, Sangkat Bavet, Krong Bavet, Svaay Rieng Cambodia AJS Electronics Ltd. 1st Floor, No. 3 Road 4, Dawei, Xinqiao, Xinqiao Community, Xinqiao Street, Baoan District, Shenzhen, Guangdong China AP Group (China) Co., Ltd. Ap Industry Garden, Quetang East District, Jinjiang, Fujian China Ability Technology (Dong Guan) Co., Ltd. Songbai Road East, Huanan Industrial Area, Liaobu Town, Donggguan, Guangdong China Anhui Goldmen Industry & Trading Co., Ltd. A-14, Zongyang Industrial Park, Tongling, Anhui China Aold Electronic Ltd. Near The Dahou Viaduct, Tianxin Industrial District, Dahou Village, Xiegang Town, Dongguan, Guangdong China Aurolite Electrical (Panyu Guangzhou) Ltd. Jinsheng Road No. 1, Jinhu Industrial Zone, Hualong, Panyu District, Guangzhou, Guangdong China Avita (Wujiang) Co., Ltd. No. 858, Jiaotong Road, Wujiang Economic Development Zone, Suzhou, Jiangsu China Bada Mechanical & Electrical Co., Ltd. No. 8 Yumeng Road, Ruian Economic Development Zone, Ruian, Zhejiang China Betec Group Ltd. -

The Spatial Analysis on Hemorrhagic Fever with Renal Syndrome in Jiangsu Province, China Based on Geographic Information System
The Spatial Analysis on Hemorrhagic Fever with Renal Syndrome in Jiangsu Province, China Based on Geographic Information System Changjun Bao2., Wanwan Liu1., Yefei Zhu2, Wendong Liu2, Jianli Hu2, Qi Liang2, Yuejia Cheng1, Ying Wu2, Rongbin Yu1, Minghao Zhou2, Hongbing Shen1, Feng Chen1, Fenyang Tang2*, Zhihang Peng1* 1 Department of Epidemiology & Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China, 2 Jiangsu Province Center for Disease Control and Prevention, Nanjing, Jiangsu, China Abstract Background: Hemorrhagic fever with renal syndrome (HFRS) is endemic in mainland China, accounting for 90% of total reported cases worldwide, and Jiangsu is one of the most severely affected provinces. In this study, the authors conducted GIS-based spatial analyses in order to determine the spatial distribution of the HFRS cases, identify key areas and explore risk factors for public health planning and resource allocation. Methods: Interpolation maps by inverse distance weighting were produced to detect the spatial distribution of HFRS cases in Jiangsu from 2001 to 2011. Spatio-temporal clustering was applied to identify clusters at the county level. Spatial correlation analysis was conducted to detect influencing factors of HFRS in Jiangsu. Results: HFRS cases in Jiangsu from 2001 to 2011 were mapped and the results suggested that cases in Jiangsu were not distributed randomly. Cases were mainly distributed in northeastern and southwestern Jiangsu, especially in Dafeng and Sihong counties. It was notable that prior to this study, Sihong county had rarely been reported as a high-risk area of HFRS. With the maximum spatial size of 50% of the total population and the maximum temporal size of 50% of the total population, spatio-temporal clustering showed that there was one most likely cluster (LLR = 624.52, P,0.0001, RR = 8.19) and one second-most likely cluster (LLR = 553.97, P,0.0001, RR = 8.25), and both of these clusters appeared from 2001 to 2004. -
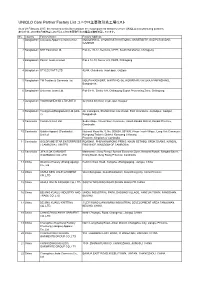
UNIQLO Core Partner Factory List ユニクロ主要取引先工場リスト
UNIQLO Core Partner Factory List ユニクロ主要取引先工場リスト As of 28 February 2017, the factories in this list constitute the major garment factories of core UNIQLO manufacturing partners. 本リストは、2017年2月末時点におけるユニクロ主要取引先の縫製工場を掲載しています。 No. Country Factory Name Factory Address 1 Bangladesh Colossus Apparel Limited unit 2 MOGORKHAL, CHOWRASTA NATIONAL UNIVERSITY, GAZIPUR SADAR, GAZIPUR 2 Bangladesh NHT Fashions Ltd. Plot no. 20-22, Sector-5, CEPZ, South Halishahar, Chittagong 3 Bangladesh Pacific Jeans Limited Plot # 14-19, Sector # 5, CEPZ, Chittagong 4 Bangladesh STYLECRAFT LTD 42/44, Chandona, Joydebpur, Gazipur 5 Bangladesh TM Textiles & Garments Ltd. MOUZA-KASHORE, WARD NO.-06, HOBIRBARI,VALUKA,MYMENSHING, Bangladesh. 6 Bangladesh Universal Jeans Ltd. Plot 09-11, Sector 6/A, Chittagong Export Processing Zone, Chittagong 7 Bangladesh YOUNGONES BD LTD UNIT-II 42 (3rd & 4th floor) Joydevpur, Gazipur 8 Bangladesh Youngones(Bangladesh) Ltd.(Unit- 24, Laxmipura, Shohid chan mia sharak, East Chandona, Joydebpur, Gazipur, 2) Bangladesh 9 Cambodia Cambo Unisoll Ltd. Seda village, Vihear Sour Commune, Ksach Kandal District, Kandal Province, Cambodia 10 Cambodia Golden Apparel (Cambodia) National Road No. 5, No. 005634, 001895, Phsar Trach Village, Long Vek Commune, Limited Kompong Tralarch District, Kompong Chhnang Province, Kingdom of Cambodia. 11 Cambodia GOLDFAME STAR ENTERPRISES ROAD#21, PHUM KAMPONG PRING, KHUM SETHBO, SROK SAANG, KANDAL ( CAMBODIA ) LIMITED PROVINCE, KINGDOM OF CAMBODIA 12 Cambodia JIFA S.OK GARMENT Manhattan ( Svay Rieng ) Special Economic Zone, National Road#, Sangkat Bavet, (CAMBODIA) CO.,LTD Krong Bavet, Svay Rieng Province, Cambodia 13 China Okamoto Hosiery (Zhangjiagang) Renmin West Road, Yangshe, Zhangjiagang, Jiangsu, China Co., Ltd 14 China ANHUI NEW JIALE GARMENT WenChangtown, XuanZhouDistrict, XuanCheng City, Anhui Province CO.,LTD 15 China ANHUI XINLIN FASHION CO.,LTD. -

Results Announcement for the Year Ended December 31, 2020
(GDR under the symbol "HTSC") RESULTS ANNOUNCEMENT FOR THE YEAR ENDED DECEMBER 31, 2020 The Board of Huatai Securities Co., Ltd. (the "Company") hereby announces the audited results of the Company and its subsidiaries for the year ended December 31, 2020. This announcement contains the full text of the annual results announcement of the Company for 2020. PUBLICATION OF THE ANNUAL RESULTS ANNOUNCEMENT AND THE ANNUAL REPORT This results announcement of the Company will be available on the website of London Stock Exchange (www.londonstockexchange.com), the website of National Storage Mechanism (data.fca.org.uk/#/nsm/nationalstoragemechanism), and the website of the Company (www.htsc.com.cn), respectively. The annual report of the Company for 2020 will be available on the website of London Stock Exchange (www.londonstockexchange.com), the website of the National Storage Mechanism (data.fca.org.uk/#/nsm/nationalstoragemechanism) and the website of the Company in due course on or before April 30, 2021. DEFINITIONS Unless the context otherwise requires, capitalized terms used in this announcement shall have the same meanings as those defined in the section headed “Definitions” in the annual report of the Company for 2020 as set out in this announcement. By order of the Board Zhang Hui Joint Company Secretary Jiangsu, the PRC, March 23, 2021 CONTENTS Important Notice ........................................................... 3 Definitions ............................................................... 6 CEO’s Letter .............................................................. 11 Company Profile ........................................................... 15 Summary of the Company’s Business ........................................... 27 Management Discussion and Analysis and Report of the Board ....................... 40 Major Events.............................................................. 112 Changes in Ordinary Shares and Shareholders .................................... 149 Directors, Supervisors, Senior Management and Staff.............................. -

Transmissibility of Hand, Foot, and Mouth Disease in 97 Counties of Jiangsu Province, China, 2015- 2020
Transmissibility of Hand, Foot, and Mouth Disease in 97 Counties of Jiangsu Province, China, 2015- 2020 Wei Zhang Xiamen University Jia Rui Xiamen University Xiaoqing Cheng Jiangsu Provincial Center for Disease Control and Prevention Bin Deng Xiamen University Hesong Zhang Xiamen University Lijing Huang Xiamen University Lexin Zhang Xiamen University Simiao Zuo Xiamen University Junru Li Xiamen University XingCheng Huang Xiamen University Yanhua Su Xiamen University Benhua Zhao Xiamen University Yan Niu Chinese Center for Disease Control and Prevention, Beijing City, People’s Republic of China Hongwei Li Xiamen University Jian-li Hu Jiangsu Provincial Center for Disease Control and Prevention Tianmu Chen ( [email protected] ) Page 1/30 Xiamen University Research Article Keywords: Hand foot mouth disease, Jiangsu Province, model, transmissibility, effective reproduction number Posted Date: July 30th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-752604/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 2/30 Abstract Background: Hand, foot, and mouth disease (HFMD) has been a serious disease burden in the Asia Pacic region represented by China, and the transmission characteristics of HFMD in regions haven’t been clear. This study calculated the transmissibility of HFMD at county levels in Jiangsu Province, China, analyzed the differences of transmissibility and explored the reasons. Methods: We built susceptible-exposed-infectious-asymptomatic-removed (SEIAR) model for seasonal characteristics of HFMD, estimated effective reproduction number (Reff) by tting the incidence of HFMD in 97 counties of Jiangsu Province from 2015 to 2020, compared incidence rate and transmissibility in different counties by non -parametric test, rapid cluster analysis and rank-sum ratio. -
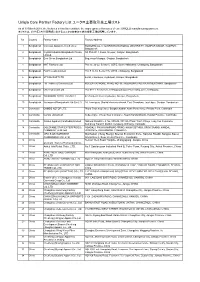
Uniqlo Core Partner Factory List ユニクロ主要取引先工場リスト
Uniqlo Core Partner Factory List ユニクロ主要取引先工場リスト As of 30 March 2018, the factories in this list constitute the major garment factories of core UNIQLO manufacturing partners. 本リストは、2018年3月30日時点におけるユニクロ主要取引先の縫製工場を掲載しています。 No. Country Factory Name Factory Address 1 Bangladesh Colossus Apparel Limited unit 2 MOGORKHAL, CHOWRASTA NATIONAL UNIVERSITY, GAZIPUR SADAR, GAZIPUR, Bangladesh 2 Bangladesh Crystal Industrial Bangladesh Private SA Plot-2013, Kewa, Sreepur, Gazipur, Bangladesh Limited. 3 Bangladesh Ever Smart Bangladesh Ltd. Begumour Mirzapur, Gazipur, Bangladesh 4 Bangladesh NHT Fashions Ltd. Plot no. 20-22, Sector-5, CEPZ, South Halishahar, Chittagong, Bangladesh 5 Bangladesh Pacific Jeans Limited Plot # 14-19, Sector # 5, CEPZ, Chittagong, Bangladesh 6 Bangladesh STYLECRAFT LTD 42/44, Chandona, Joydebpur, Gazipur, Bangladesh 7 Bangladesh TM Textiles & Garments Ltd. MOUZA-KASHORE, WARD NO.-06, HOBIRBARI,VALUKA,MYMENSHING, Bangladesh 8 Bangladesh Universal Jeans Ltd. Plot 09-11, Sector 6/A, Chittagong Export Processing Zone, Chittagong 9 Bangladesh YOUNGONES BD LTD UNIT-II 42 (3rd & 4th floor) Joydevpur, Gazipur, Bangladesh 10 Bangladesh Youngones(Bangladesh) Ltd.(Unit-2) 24, Laxmipura, Shohid chan mia sharak, East Chandona, Joydebpur, Gazipur, Bangladesh 11 Cambodia CAMBO KOTOP LTD. Phum Trapeang Chrey, Sangkat Kakab, Khan Porsenchey, Phnom Penh, Cambodia. 12 Cambodia Cambo Unisoll Ltd. Seda village, Vihear Sour Commune, Ksach Kandal District, Kandal Province, Cambodia 13 Cambodia Golden Apparel (Cambodia) Limited National Road No. -

Spatial and Temporal Evaluation of Ecological Footprint Intensity of Jiangsu Province at the County-Level Scale
International Journal of Environmental Research and Public Health Article Spatial and Temporal Evaluation of Ecological Footprint Intensity of Jiangsu Province at the County-Level Scale Decun Wu 1 and Jinping Liu 2,* 1 School of Public Administration and Sociology, Jiangsu Normal University, Xuzhou 221116, China; [email protected] 2 School of Economics and Management, China University of Mining and Technology, Xuzhou 221116, China * Correspondence: [email protected] Received: 10 September 2020; Accepted: 23 October 2020; Published: 26 October 2020 Abstract: Due to the high ecological pressure that exists in the process of rapid economic development in Jiangsu Province, it is necessary to evaluate its ecological footprint intensity (EFI). This article focuses on ecological footprint intensity analysis at the county scale. We used county-level data to evaluate the spatial distributions and temporal trends of the ecological footprint intensity in Jiangsu’s counties from 1995 to 2015. The temporal trends of counties are divided into five types: linear declining type, N-shape type, inverted-N type, U-shape type and inverted-U shape type. It was discovered that the proportions of the carbon footprint intensity were maintained or increased in most counties. Exploratory spatial data analysis shows that there was a certain regularity of the EFI spatial distributions, i.e., a gradient decrease from north to south, and there was a decline in the spatial heterogeneity of EFI in Jiangsu’s counties over time. The global Moran’s index (Moran’s I) and local spatial association index (LISA) are used to analyze both the global and local spatial correlation of EFIs among counties of Jiangsu Province. -
Cambridge University Press 978-1-107-16629-5 — Incentivized Development in China David J
Cambridge University Press 978-1-107-16629-5 — Incentivized Development in China David J. Bulman Index More Information 267 Index accountability corruption, 1, 2, 5, 7, 7n. 12, 9, 10, 12, downward accountability, 6, 10, 14, 14n. 15n. 26, 25, 27, 77, 77n. 4, 78, 79, 23, 79, 83, 106, 116, 153, 228, 230n. 79n. 13, 80, 80n. 16, 82, 83, 85, 86, 3, 231, 232 86n. 27, 86n. 28, 87, 87n. 31, 87n. 32, horizontal accountability, 84, 116, 117 88, 89, 90, 90n. 36, 91, 92, 92n. 37, upward accountability, 10, 16, 83, 92n. 38, 92n. 39, 93, 94, 95, 95n. 43, 116, 145n. 51, 153, 225, 227, 230, 96t.3.1., 102n. 50, 103n. 54, 106, 110, 230n. 3 111, 113, 114, 116, 127, 141, 142, Agricultural Bank of China, 49 145, 145n. 51, 149, 149n. 57, 157, Allison, Graham, 201 158n. 15, 161n. 18, 164, 171, 197, Asian Financial Crisis, 7, 119 199n. 11, 216, 224, 230, 230n. 3 attapulgite, 27, 55, 148 county branding, 136– 39 authoritarian fragmentation, 3 Deng Xiaoping, 31, 210 Bank of China, 49 development zones, 14, 27, 40n. 19,43– 45, Beijing, 18, 20n. 35, 109, 130n. 26, 138, 44n. 35, 51, 73, 98, 99, 100, 101n. 47, 139, 141, 162, 164, 185, 193, 198, 104, 110, 134, 136n. 36, 220 198n. 10, 200, 204n. 22, 205t.6.1., developmental state, 4, 7, 11, 12, 85, 99n. 46 212n. 33, 233 Dongxiang Autonomous County, 52 Bengbu City, Anhui, 26, 149, 165, 185, Dongyang City, Zhejiang, 200 233 dujin, 179, 184, 187, 188– 91, 191n. -

Subsidy Programs
SUBSIDIES REQUEST FROM THE UNITED STATES TO CHINA PURSUANT TO ARTICLE 25.10 OF THE AGREEMENT The following communication, dated 15 April 2016, is being circulated at the request of the Delegation of the United States. _______________ In the report that it prepared for China's most recent Trade Policy Review, held in July 2014, the Secretariat included information that it had uncovered on 30 support programs for China's fisheries sector.1 The Secretariat's Report noted that China had not notified any of these support programs to the Committee on Subsidies and Countervailing Measures (Committee) and that China could not verify any of the information provided to it by the Secretariat. During the ensuing Trade Policy Review meetings, China responded to Members' questions about these support programs by stating that it needed more time to identify and verify information regarding these support programs. In April of last year, the United States submitted questions to China with respect to these programs and other fishery support measures the United States had uncovered through its own efforts.2 Once again, despite its obligation under Article 25.9 of the Agreement to provide answers "as quickly as possible and in a comprehensive manner", China to date has not meaningfully responded to the United States' request and has refused repeated requests to meet bilaterally to discuss the issue, and the issue of subsidy notifications more generally. In November of last year, China submitted its latest subsidy notification covering the period 2009 through 2014.3 In this notification, China did not include: (1) any of the fishery subsidy programs identified in China's 2014 TPR report, (2) any of the additional measures identified by the United States in its Article 25.8 submission or (3) any of the fisheries subsidies measures identified in the 2014 Article 25.10 submission of the United States4. -

World Bank Document
Public Disclosure Authorized ENVIRONMENTAL IMPACT ASSESSMENT OF THE YANGCHENG TO HlJAIYIN 500 kV POWER TRANSMISSION l'ROJECT Public Disclosure Authorized V.i.. 3 Public Disclosure Authorized Jiangsu Provincial Electric Power Company No. 20 West Beijing Road Nanjing, Jiangsu Province People's Republic of China October 1997 Public Disclosure Authorized Table of Contents Pages 0. Executive summary 0-0 1. Introduction and background 1-1 I.1 The necessity of 500kV power transmission project construction 1-2 41.2The basic of environmental impact assessment for 1-2 500kV power transmission project 2. Project description 2-1 2.1 The route and composition of power transmission lines 2-1 2.2 Svitchyard. substation 2-2 2.3 power transmission line 2-2 2.4 Yellow river crossing 2-4 3. Environmental background status 3 3.1INatural environment 3-1 3.2 Ecological environment 3-3 3.3 Noise level 3-4 3.4 Electric field level 3-5 3.5 Social economic environment and living quality environment 3-5 4. Construction periiod environmental impact 4-1 4.1 Environmental impact identification 4-1 4.2 The impact on natural environment 4-1 4.3 Impact on ecological environment 4-4 4.4 Impact on social environment 4-6 4.5 Impact on living quality environment 4-8 5. Operation period environmental impact 5-1 5.1 Environmental impact identification 5-1 5.2 Impact on natural environment 5-1 5.3 Impact on ecological environment 5-3 5.4 Impact on social environment 5-5 5.5 Impact on living environment 5-8 6. -

12344 15926 Lidl Transparentnost V Nepotravinovom
Zoznam hlavných výrobných miest pre vlastné značky z hardvérového sortimentu Výrobca Adresa Krajina Allplast Bangladesh Ltd. Mulgaon, Kaliganj, Gazipur, Rfl Industrial Park Rip, Bangladesh Mulgaon, Sandanpara, Kaligonj, Gazipur, Dhaka Bengal Plastics Ltd. (Unit - 3) Yearpur, Zirabo Bazar, Savar, Dhaka Bangladesh Durable Plastic Ltd. Mulgaon, Kaligonj, Gazipur, Dhaka Bangladesh HKD International (Cepz) Ltd. Plot # 49-52, Sector # 8, Cepz, Chittagong Bangladesh Lhotse (Bd) Ltd. Plot No. 60 & 61, Sector -3, Karnaphuli Export Processing Bangladesh Zone, North Potenga, Chittagong Plastofl ex Doo Branilaca Grada Bb, Gračanica, Federacija Bosne I H Bosnia-Herz. ASF Sporting Goods Co., Ltd. Km 38.5, National Road No. 3, Thlork Village, Chonrok Cambodia Commune, Konrrg Pisey, Kampong Spueu Powerjet Home Product Manhattan (Svay Rieng) Special Economic Zone, National Cambodia (Cambodia) Co., Ltd. Road 1, Sangkat Bavet, Krong Bavet, Svaay Rieng AJS Electronics Ltd. 1st Floor, No. 3 Road 4, Dawei, Xinqiao, Xinqiao China Community, Xinqiao Street, Baoan District, Shenzhen, Guangdong AP Group (China) Co., Ltd. Ap Industry Garden, Quetang East District, Jinjiang, Fujian China Ability Technology (Dong Guan) Songbai Road East, Huanan Industrial Area, Liaobu Town, China Co., Ltd. Donggguan, Guangdong Anhui Goldmen Industry & A-14, Zongyang Industrial Park, Tongling, Anhui China Trading Co., Ltd. Aold Electronic Ltd. Near The Dahou Viaduct, Tianxin Industrial District, Dahou China Village, Xiegang Town, Dongguan, Guangdong Aurolite Electrical (Panyu Jinsheng Road No. 1, Jinhu Industrial Zone, Hualong, Panyu China Guangzhou) Ltd. District, Guangzhou, Guangdong Avita (Wujiang) Co., Ltd. No. 858, Jiaotong Road, Wujiang Economic Development China Zone, Suzhou, Jiangsu Bada Mechanical & Electrical Co., No. 8 Yumeng Road, Ruian Economic Development Zone, China Ltd. -

Research on the Translation of Liquor Brands in Huaian City
ISSN 1799-2591 Theory and Practice in Language Studies, Vol. 6, No. 2, pp. 352-356, February 2016 DOI: http://dx.doi.org/10.17507/tpls.0602.17 Research on the Translation of Liquor Brands in Huaian City Xiangfeng Guan Faculty of Foreign Languages of Huaiyin Institute of Technology, Huaian, Jiangsu, China Abstract—In this paper, combining with the British translation theorist, Peter Newmark, on the classification of text types, namely, the expressive text type, the informative text type and calling attention text type, the author puts forward the latter two kinds of text types are fit for the translation of liquor brands. In the process of translation of liquor brands, on the premise of adhering to the principles of commodity brand translation and Cuisine Translation Principles, we should also consider the local liquor culture characteristics. At the same time, we should adopt the legal requirements of developed countries to add warnings of excessive drinking is harmful and no drunk driving to liquor product logos. Index Terms—Huai liquor, the principles of liquor brands translation, Chinese pinyin domestication translation strategy, warnings I. INTRODUCTION Chinese liquor culture has a long history, its fame does not only come from its extensiveness, profoundness, vast territory and variety, but also from its unique fragrance, mysterious brewing process and production of liquor, which enjoys a good reputation both at home and abroad. In terms of the types, there are five main categories: liquor, rice wine, beer, fruit wine and mixed wine. If we classify liquor according to their different flavors, there are maotai-flavor, luzhou-flavor, light fragrant flavor, rice fragrant flavor, phoenix fragrant flavor, sesame fragrant flavor , fermented bean fragrant flavor and special fragrant flavor, altogether 8 categories (Gao, 2002).