Improving the Management of Patients with Hearing Loss by the Implementation of an NGS Panel in Clinical Practice
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Genetic Analysis of Retinopathy in Type 1 Diabetes
Genetic Analysis of Retinopathy in Type 1 Diabetes by Sayed Mohsen Hosseini A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Institute of Medical Science University of Toronto © Copyright by S. Mohsen Hosseini 2014 Genetic Analysis of Retinopathy in Type 1 Diabetes Sayed Mohsen Hosseini Doctor of Philosophy Institute of Medical Science University of Toronto 2014 Abstract Diabetic retinopathy (DR) is a leading cause of blindness worldwide. Several lines of evidence suggest a genetic contribution to the risk of DR; however, no genetic variant has shown convincing association with DR in genome-wide association studies (GWAS). To identify common polymorphisms associated with DR, meta-GWAS were performed in three type 1 diabetes cohorts of White subjects: Diabetes Complications and Control Trial (DCCT, n=1304), Wisconsin Epidemiologic Study of Diabetic Retinopathy (WESDR, n=603) and Renin-Angiotensin System Study (RASS, n=239). Severe (SDR) and mild (MDR) retinopathy outcomes were defined based on repeated fundus photographs in each study graded for retinopathy severity on the Early Treatment Diabetic Retinopathy Study (ETDRS) scale. Multivariable models accounted for glycemia (measured by A1C), diabetes duration and other relevant covariates in the association analyses of additive genotypes with SDR and MDR. Fixed-effects meta- analysis was used to combine the results of GWAS performed separately in WESDR, ii RASS and subgroups of DCCT, defined by cohort and treatment group. Top association signals were prioritized for replication, based on previous supporting knowledge from the literature, followed by replication in three independent white T1D studies: Genesis-GeneDiab (n=502), Steno (n=936) and FinnDiane (n=2194). -

Missense Variant in LOXHD1 Is Associated with Canine Nonsyndromic Hearing Loss
Missense Variant in LOXHD1 is Associated With Canine Nonsyndromic Hearing Loss Marjo K Hytönen University of Helsinki: Helsingin Yliopisto Julia E Niskanen University of Helsinki: Helsingin Yliopisto Meharji Arumilli University of Helsinki: Helsingin Yliopisto Casey A Knox Wisdom Health Jonas Donner Genoscoper Laboratories Hannes Lohi ( hannes.lohi@helsinki. ) Helsingin Yliopisto Laaketieteellinen tiedekunta https://orcid.org/0000-0003-1087-5532 Research Article Keywords: dog, hearing, hearing loss, deafness, Rottweiler, stereocilia, PLAT Posted Date: March 16th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-288479/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/16 Abstract Hearing loss is a common sensory decit both in humans and dogs. In canines the genetic basis is largely unknown, as genetic variants have only been identied for a syndromic form of hearing impairment. We observed a congenital or early-onset sensorineural hearing loss in a Rottweiler litter. Assuming an autosomal recessive inheritance, we used a combined approach of homozygosity mapping and genome sequencing to dissect the genetic background of the disorder. We identied a fully segregating missense variant in LOXHD1, a gene that is known to be essential for cochlear hair cell function and associated with nonsyndromic hearing loss in humans and mice. The canine LOXHD1 variant was specic to the Rottweiler breed in our study cohorts of pure-bred dogs. However, it also was present in mixed-breed dogs, of which the majority showed Rottweiler ancestry. Low allele frequencies in these populations, 2.6 % and 0.04 %, respectively, indicate a rare variant. -

Greg's Awesome Thesis
Analysis of alignment error and sitewise constraint in mammalian comparative genomics Gregory Jordan European Bioinformatics Institute University of Cambridge A dissertation submitted for the degree of Doctor of Philosophy November 30, 2011 To my parents, who kept us thinking and playing This dissertation is the result of my own work and includes nothing which is the out- come of work done in collaboration except where specifically indicated in the text and acknowledgements. This dissertation is not substantially the same as any I have submitted for a degree, diploma or other qualification at any other university, and no part has already been, or is currently being submitted for any degree, diploma or other qualification. This dissertation does not exceed the specified length limit of 60,000 words as defined by the Biology Degree Committee. November 30, 2011 Gregory Jordan ii Analysis of alignment error and sitewise constraint in mammalian comparative genomics Summary Gregory Jordan November 30, 2011 Darwin College Insight into the evolution of protein-coding genes can be gained from the use of phylogenetic codon models. Recently sequenced mammalian genomes and powerful analysis methods developed over the past decade provide the potential to globally measure the impact of natural selection on pro- tein sequences at a fine scale. The detection of positive selection in particular is of great interest, with relevance to the study of host-parasite conflicts, immune system evolution and adaptive dif- ferences between species. This thesis examines the performance of methods for detecting positive selection first with a series of simulation experiments, and then with two empirical studies in mammals and primates. -

Genetic Mutations and Molecular Mechanisms of Fuchs Endothelial
Liu et al. Eye and Vision (2021) 8:24 https://doi.org/10.1186/s40662-021-00246-2 REVIEW Open Access Genetic mutations and molecular mechanisms of Fuchs endothelial corneal dystrophy Xuerui Liu1†, Tao Zheng1†, Chuchu Zhao1, Yi Zhang1, Hanruo Liu2, Liyuan Wang1* and Ping Liu1* Abstract Background: Fuchs endothelial corneal dystrophy is a hereditary disease and the most frequent cause of corneal transplantation in the worldwide. Its main clinical signs are an accelerated decrease in the number of endothelial cells, thickening of Descemet’s membrane and formation of guttae in the extracellular matrix. The cornea’s ability to maintain stromal dehydration is impaired, causing painful epithelial bullae and loss of vision at the point when the amount of corneal endothelial cells cannot be compensated. At present, apart from corneal transplantation, there is no other effective treatment that prevents blindness. Main text: In this review, we first summarized the mutations of COL8A2, TCF4, TCF8, SLC4A11 and AGBL1 genes in Fuchs endothelial corneal dystrophy. The molecular mechanisms associated with Fuchs endothelial corneal dystrophy, such as endoplasmic reticulum stress and unfolded protein response pathway, oxidative stress, mitochondrial dysregulation pathway, apoptosis pathway, mitophagy, epithelial-mesenchymal transition pathway, RNA toxicity and repeat-associated non-ATG translation, and other pathogenesis, were then explored. Finally, we discussed several potential treatments related to the pathogenesis of Fuchs endothelial corneal dystrophy, which may be the focus of future research. Conclusions: The pathogenesis of Fuchs endothelial corneal dystrophy is very complicated. Currently, corneal transplantation is an important method in the treatment of Fuchs endothelial corneal dystrophy. It is necessary to continuously explore the pathogenesis of Fuchs endothelial corneal dystrophy and establish the scientific foundations for the development of next-generation corneal therapeutics. -
Supplementary Figure 1
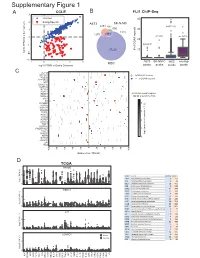
6XSSOHPHQWDU\)LJXUH CCLE B FLI1 ChIP-Seq A 3 FRPPRQ 40 (ZLQJ6SHFLILF 2 A673 6.10& 477 Q 690 30 0 UHSHDWV Q Q A 20 í Q 57226 RI**$ ORJ)3.0LQRWKHUFDQFHUV í í A673 6.10& 06& RYHUODS í 0 23 06& ORJ)3.0LQ(ZLQJ6DUFRPD SHDNV SHDNV SHDNV SHDNV & ./) 67($3 (:6)/,ERXQG 3'(% ! **$AUHSHDWV 3335$ ABI3 '1$-& '863 13<5 .&1( 5%0 DOO**$AUHSHDWUHJLRQV 1.; NEDURXQGWKH766 9$9 ;* .&1$% 4 .&1$ 51) 0<20 3 355/ 355T4 &'$ $'5% 2 '&'& 711, 8*7$ 3+263+2 )(=) *1*7 ORJ QXPEHURIUHSHDWV A571 0 13<5 /2;+' /,3, 5$; 0 40 60 20 GLVWDQFHIURP766 NE D TCGA 35$0( 5 code cancer normal tumor LAML Acute Myeloid Leukemia 0 151 ORJ )3.0 ACC AdrenocoƌƟcal carcinoma 0 79 0 BLCA Bladder Urothelial Carcinoma 19 411 LGG Brain Lower Grade Glioma 0 529 COAD Colon adenocarcinoma 41 464 5%0 ESCA Esophageal carcinoma 11 162 GBM Glioblastoma muůƟforme 5 168 5 KICH Kidney Chromophobe 24 65 KIRP Kidney renal papillary cell carcinoma 32 289 LIHC Liver hepatocellular carcinoma 50 374 ORJ )3.0 LUAD Lung adenocarcinoma 59 526 0 LUSC Lung squamous cell carcinoma 49 501 DLBC Lymphoid Neoplasm Diīuse Large B-cell Lym 042 /,3, MESO Mesothelioma 0 1 OV Ovarian serous cystadenocarcinoma 0 379 PAAD PancreĂƟc adenocarcinoma 4 178 5 PCPG Pheochromocytoma and Paraganglioma 3 150 PRAD Prostate adenocarcinoma 52 499 READ Rectum adenocarcinoma 1 6 ORJ )3.0 SARC Sarcoma 0 35 0 SKCM Skin Cutaneous Melanoma 1 471 STAD Stomach adenocarcinoma 32 375 /2;+' WXPRU TGCT dĞƐƟĐƵůĂƌ'ĞƌŵĞůůdƵŵŽƌƐ 0156 THCA Thyroid carcinoma 58 510 QRUPDO UCS Uterine Carcinosarcoma 0 56 5 ORJ )3.0 0 / / 29 $&& 8&6 /** *%0 -

Variation in Protein Coding Genes Identifies Information Flow
bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 1 1 2 3 4 5 Variation in protein coding genes identifies information flow as a contributor to 6 animal complexity 7 8 Jack Dean, Daniela Lopes Cardoso and Colin Sharpe* 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 Institute of Biological and Biomedical Sciences 25 School of Biological Science 26 University of Portsmouth, 27 Portsmouth, UK 28 PO16 7YH 29 30 * Author for correspondence 31 [email protected] 32 33 Orcid numbers: 34 DLC: 0000-0003-2683-1745 35 CS: 0000-0002-5022-0840 36 37 38 39 40 41 42 43 44 45 46 47 48 49 Abstract bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 2 1 Across the metazoans there is a trend towards greater organismal complexity. How 2 complexity is generated, however, is uncertain. Since C.elegans and humans have 3 approximately the same number of genes, the explanation will depend on how genes are 4 used, rather than their absolute number. -

Mutational Spectrum and Clinical Features of Patients with LOXHD1 Variants Identified in an 8074 Hearing Loss Patient Cohort
G C A T T A C G G C A T genes Article Mutational Spectrum and Clinical Features of Patients with LOXHD1 Variants Identified in an 8074 Hearing Loss Patient Cohort Karuna Maekawa 1, Shin-ya Nishio 1,2 , Satoko Abe 3, Shin-ichi Goto 4, Yohei Honkura 5, Satoshi Iwasaki 6, Yukihiko Kanda 7, Yumiko Kobayashi 8, Shin-ichiro Oka 6, Mayuri Okami 9, Chie Oshikawa 10, Naoko Sakuma 11, Hajime Sano 12 , Masayuki Shirakura 5, Natsumi Uehara 13 and Shin-ichi Usami 1,2,* 1 Department of Otorhinolaryngology, Shinshu University School of Medicine, 3-1-1 Asahi, Matsumoto, Nagano 390-8621, Japan; [email protected] (K.M.); [email protected] (S.-y.N.) 2 Department of Hearing Implant Sciences, Shinshu University School of Medicine, 3-1-1 Asahi, Matsumoto, Nagano 390-8621, Japan 3 Department of Otorhinolaryngology, Toranomon Hosipital, 2-2-2 Toranomon, Minato-ku, Tokyo 105-8470, Japan; [email protected] 4 Department of Otorhinolaryngology, Hirosaki University School of Medicine, 53 Honcho, Hirosaki, Aomori 036-8203, Japan; [email protected] 5 Department of Otolaryngology-Head and Neck Surgery, Tohoku University School of Medicine, 1-1 Seiryomachi, Aoba-ku, Sendai, Miyagi 980-0872, Japan; [email protected] (Y.H.); [email protected] (M.S.) 6 Department of Otorhinolaryngology, International University of Health and Welfare, Mita Hospital, 1-4-3 Mita, Minato-ku, Tokyo 108-8329, Japan; [email protected] (S.I.); [email protected] (S.-i.O.) 7 Kanda ENT Clinic, Nagasaki Bell Hearing Center, 4-25 Wakakusa-cho, Nagasaki, -

Mutations in LOXHD1, an Evolutionarily Conserved Stereociliary Protein, Disrupt Hair Cell Function in Mice and Cause Progressive Hearing Loss in Humans
ARTICLE Mutations in LOXHD1, an Evolutionarily Conserved Stereociliary Protein, Disrupt Hair Cell Function in Mice and Cause Progressive Hearing Loss in Humans Nicolas Grillet,1,11 Martin Schwander,1,11 Michael S. Hildebrand,2 Anna Sczaniecka,1 Anand Kolatkar,1 Janice Velasco,3 Jennifer A. Webster,4 Kimia Kahrizi,5 Hossein Najmabadi,5 William J. Kimberling,6 Dietrich Stephan,3,7,8 Melanie Bahlo,9 Tim Wiltshire,10 Lisa M. Tarantino,10 Peter Kuhn,1 Richard J.H. Smith,2 and Ulrich Mu¨ller1,* Hearing loss is the most common form of sensory impairment in humans and is frequently progressive in nature. Here we link a previ- ously uncharacterized gene to hearing impairment in mice and humans. We show that hearing loss in the ethylnitrosourea (ENU)- induced samba mouse line is caused by a mutation in Loxhd1. LOXHD1 consists entirely of PLAT (polycystin/lipoxygenase/a-toxin) domains and is expressed along the membrane of mature hair cell stereocilia. Stereociliary development is unaffected in samba mice, but hair cell function is perturbed and hair cells eventually degenerate. Based on the studies in mice, we screened DNA from human families segregating deafness and identified a mutation in LOXHD1, which causes DFNB77, a progressive form of autosomal-recessive nonsyndromic hearing loss (ARNSHL). LOXHD1, MYO3a, and PJVK are the only human genes to date linked to progressive ARNSHL. These three genes are required for hair cell function, suggesting that age-dependent hair cell failure is a common mechanism for progres- sive ARNSHL. Introduction orthologous genes in these species causing comparable phenotypes.5–9 However, examples of genetic mutations Hearing impairment in humans is frequently genetic in that lead to progressive autosomal-recessive nonsyn- origin and progressive in nature. -
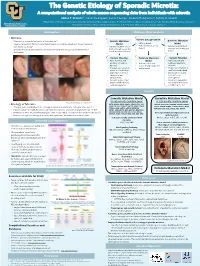
Example of a Scientific Poster
The Genetic Etiology of Sporadic Microtia: A computational analysis of whole exome sequencing data from individuals with microtia Abbas T. Shaikh1,*, Iván E. Rodríguez2, Connor Lester3, Frederic Deleyiannis4, Tamim H. Shaikh1 1Department of Pediatrics, University of Colorado, Anschutz Medical Campus, Aurora, CO; 2Department of Surgery, University of Colorado, Anschutz Medical Campus, Aurora, CO; University of Colorado Denver 3 4 Anschutz Medical Campus Georgetown University School of Medicine, Washington, D.C.; UCHealth Medical Group, Colorado Springs, CO Introduction Methods: Data Analysis • Microtia Variant Categorization • Microtia is a congenital deformity of the outer ear[1] Somatic Mutation Germline Mutation • • Severity ranges from mild structural deformations to complete absence of the auricle (outer Model • SNVs or Indels Model • high, moderate, or low • ear), known as anotia[1] • Variants found in one or • high, moderate, or low • Variants found in both impact affected and unaffected • Sporadic microtia is an isolated occurrence of microtia with no associated syndromes or both affected tissues but affected and unaffected tissues deformities not in unaffected tissues Variant Filtering Common Mutation Variant Filtering • Gene function and Model • Gene function and pathway relevant to • Variants consistent pathway relevant to microtia across multiple patients microtia • Frequency of variant in with microtia • Frequency of variant in unaffected individuals unaffected individuals • Expression in tissues • Expression in tissues relevant to ear relevant to ear development development • Variants in genes that • Variants in genes that matched with microtia matched with microtia candidate gene list candidate gene list (204) Results Somatic Mutation Model Germline Mutation Model 37 high-priority candidate genes: 18 high-priority candidate genes: Image taken from Luquetti et al. -

Mutations in LOXHD1, a Recessive-Deafness Locus, Cause Dominant Late-Onset Fuchs Corneal Dystrophy
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector REPORT Mutations in LOXHD1, a Recessive-Deafness Locus, Cause Dominant Late-Onset Fuchs Corneal Dystrophy S. Amer Riazuddin,1 David S. Parker,2 Elyse J. McGlumphy,1 Edwin C. Oh,2 Benjamin W. Iliff,1 Thore Schmedt,3 Ula Jurkunas,3 Robert Schleif,4 Nicholas Katsanis,2 and John D. Gottsch1,* Fuchs corneal dystrophy (FCD) is a genetic disorder of the corneal endothelium and is the most common cause of corneal transplanta- tion in the United States. Previously, we mapped a late-onset FCD locus, FCD2, on chromosome 18q. Here, we present next-generation sequencing of all coding exons in the FCD2 critical interval in a multigenerational pedigree in which FCD segregates as an autosomal- dominant trait. We identified a missense change in LOXHD1, a gene causing progressive hearing loss in humans, as the sole variant capable of explaining the phenotype in this pedigree. We observed LOXHD1 mRNA in cultured human corneal endothelial cells, whereas antibody staining of both human and mouse corneas showed staining in the corneal epithelium and endothelium. Corneal sections of the original proband were stained for LOXHD1 and demonstrated a distinct increase in antibody punctate staining in the endothelium and Descemet membrane; punctate staining was absent from both normal corneas and FCD corneas negative for causal LOXHD1 mutations. Subsequent interrogation of a cohort of >200 sporadic affected individuals identified another 15 heterozygous missense mutations that were absent from >800 control chromosomes. Furthermore, in silico analyses predicted that these mutations reside on the surface of the protein and are likely to affect the protein’s interface and protein-protein interactions. -
Target Sequencing of 307 Deafness Genes Identifies Candidate Genes Implicated in Microtia
www.impactjournals.com/oncotarget/ Oncotarget, 2017, Vol. 8, (No. 38), pp: 63324-63332 Research Paper Target sequencing of 307 deafness genes identifies candidate genes implicated in microtia Pu Wang1, Xinmiao Fan1, Yibei Wang1, Yue Fan1, Yaping Liu2, Shuyang Zhang3 and Xiaowei Chen1 1Department of Otolaryngology, Peking Union Medical College Hospital, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China 2Department of Medical Genetics, School of Basic Medicine, Peking Union Medical College, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China 3Department of Cardiology, Peking Union Medical College Hospital, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China Correspondence to: Xiaowei Chen, email: [email protected] Shuyang Zhang, email: [email protected] Keywords: microtia, deafness genes, next-generation sequencing, SKAT Received: April 23, 2017 Accepted: May 29, 2017 Published: June 28, 2017 Copyright: Wang et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Microtia is a congenital malformation of the external ear caused by genetic and/or environmental factors. However, no causal genetic mutations have been identified in isolated microtia patients. In this study, we utilized targeted genomic capturing combined with next-generation sequencing to screen for mutations in 307 deafness genes in 32 microtia patients. Forty-two rare heterozygous mutations in 25 genes, including 22 novel mutations in 24 isolated unilateral microtia cases were identified. Pathway analysis found five pathways especially focal adhesion pathway and ECM-receptor interaction pathway were significantly associated with microtia. -
Genetics of Hearing Loss in the Arab Population of Northern Israel
European Journal of Human Genetics (2018) 26:1840–1847 https://doi.org/10.1038/s41431-018-0218-z ARTICLE Genetics of hearing loss in the Arab population of Northern Israel 1,2,3 3 4 3 1 1 Nada Danial-Farran ● Zippora Brownstein ● Suleyman Gulsuner ● Luna Tammer ● Morad Khayat ● Ola Aleme ● 1 1 4 3 4 3 Elena Chervinsky ● Olfat Aboleile Zoubi ● Tom Walsh ● Gil Ast ● Mary-Claire King ● Karen B. Avraham ● Stavit A. Shalev1,2 Received: 21 February 2018 / Revised: 18 June 2018 / Accepted: 26 June 2018 / Published online: 23 August 2018 © The Author(s) 2018. This article is published with open access Abstract For multiple generations, much of the Arab population of Northern Israel has lived in communities with consanguineous marriages and large families. These communities have been particularly cooperative and informative for understanding the genetics of recessive traits. We studied the genetics of hearing loss in this population, evaluating 168 families from 46 different villages. All families were screened for founder variants by Sanger sequencing and 13 families were further evaluated by sequencing all known genes for hearing loss using our targeted gene panel HEar-Seq. Deafness in 34 of 168 families (20%) was explained by founder variants in GJB2, SLC26A4,orOTOF. In 6 of 13 families (46%) evaluated using HEar-Seq, deafness was explained by damaging alleles of SLC26A4, MYO15A, OTOG, LOXHD1, and TBC1D24. In some 1234567890();,: 1234567890();,: genes critical to hearing, it is particularly difficult to interpret variants that might affect splicing, because the genes are not expressed in accessible tissue. To address this problem for possible splice-altering variants of MYO15A, we evaluated minigenes transfected into HEK293 cells.