Supplementary Data Patients and Methods
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Analysis of Retinopathy in Type 1 Diabetes
Genetic Analysis of Retinopathy in Type 1 Diabetes by Sayed Mohsen Hosseini A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Institute of Medical Science University of Toronto © Copyright by S. Mohsen Hosseini 2014 Genetic Analysis of Retinopathy in Type 1 Diabetes Sayed Mohsen Hosseini Doctor of Philosophy Institute of Medical Science University of Toronto 2014 Abstract Diabetic retinopathy (DR) is a leading cause of blindness worldwide. Several lines of evidence suggest a genetic contribution to the risk of DR; however, no genetic variant has shown convincing association with DR in genome-wide association studies (GWAS). To identify common polymorphisms associated with DR, meta-GWAS were performed in three type 1 diabetes cohorts of White subjects: Diabetes Complications and Control Trial (DCCT, n=1304), Wisconsin Epidemiologic Study of Diabetic Retinopathy (WESDR, n=603) and Renin-Angiotensin System Study (RASS, n=239). Severe (SDR) and mild (MDR) retinopathy outcomes were defined based on repeated fundus photographs in each study graded for retinopathy severity on the Early Treatment Diabetic Retinopathy Study (ETDRS) scale. Multivariable models accounted for glycemia (measured by A1C), diabetes duration and other relevant covariates in the association analyses of additive genotypes with SDR and MDR. Fixed-effects meta- analysis was used to combine the results of GWAS performed separately in WESDR, ii RASS and subgroups of DCCT, defined by cohort and treatment group. Top association signals were prioritized for replication, based on previous supporting knowledge from the literature, followed by replication in three independent white T1D studies: Genesis-GeneDiab (n=502), Steno (n=936) and FinnDiane (n=2194). -

Missense Variant in LOXHD1 Is Associated with Canine Nonsyndromic Hearing Loss
Missense Variant in LOXHD1 is Associated With Canine Nonsyndromic Hearing Loss Marjo K Hytönen University of Helsinki: Helsingin Yliopisto Julia E Niskanen University of Helsinki: Helsingin Yliopisto Meharji Arumilli University of Helsinki: Helsingin Yliopisto Casey A Knox Wisdom Health Jonas Donner Genoscoper Laboratories Hannes Lohi ( hannes.lohi@helsinki. ) Helsingin Yliopisto Laaketieteellinen tiedekunta https://orcid.org/0000-0003-1087-5532 Research Article Keywords: dog, hearing, hearing loss, deafness, Rottweiler, stereocilia, PLAT Posted Date: March 16th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-288479/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/16 Abstract Hearing loss is a common sensory decit both in humans and dogs. In canines the genetic basis is largely unknown, as genetic variants have only been identied for a syndromic form of hearing impairment. We observed a congenital or early-onset sensorineural hearing loss in a Rottweiler litter. Assuming an autosomal recessive inheritance, we used a combined approach of homozygosity mapping and genome sequencing to dissect the genetic background of the disorder. We identied a fully segregating missense variant in LOXHD1, a gene that is known to be essential for cochlear hair cell function and associated with nonsyndromic hearing loss in humans and mice. The canine LOXHD1 variant was specic to the Rottweiler breed in our study cohorts of pure-bred dogs. However, it also was present in mixed-breed dogs, of which the majority showed Rottweiler ancestry. Low allele frequencies in these populations, 2.6 % and 0.04 %, respectively, indicate a rare variant. -

Transgelin Interacts with PARP1 and Affects Rho Signaling Pathway in Human Colon Cancer Cells
Transgelin interacts with PARP1 and affects Rho signaling pathway in human colon cancer cells Zhen-xian Lew Guangzhou Concord Cancer Center Hui-min Zhou First Aliated Hospital of Guangdong Pharmaceutical College Yuan-yuan Fang Tongling Peoples's Hospital Zhen Ye Sun Yat-sen Memorial Hospital Wa Zhong Sun Yat-sen Memorial Hospital Xin-yi Yang The Seventh Aliated Hospital Sun Yat-sen University Zhong Yu Sun Yat-sen Memorial Hospital of Sun Yat-sen University Dan-yu Chen Sun Yat-sen Memorial Hospital Si-min Luo Sun Yat-sen Memorial Hospital Li-fei Chen Sun Yat-sen Memorial Hospital Ying Lin ( [email protected] ) Sun Yat-sen Memorial Hospital https://orcid.org/0000-0003-2416-2154 Primary research Keywords: Transgelin, PARP1, Colon Cancer, Rho Signaling, Bioinformatics Posted Date: July 1st, 2020 DOI: https://doi.org/10.21203/rs.3.rs-16964/v2 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/22 Version of Record: A version of this preprint was published on August 3rd, 2020. See the published version at https://doi.org/10.1186/s12935-020-01461-y. Page 2/22 Abstract Background: Transgelin, an actin-binding protein, is associated with the cytoskeleton remodeling. Our previous studies found that transgelin was up-regulated in node-positive colorectal cancer versus in node- negative disease. Over-expression of TAGLN affected the expression of 256 downstream transcripts and increased the metastatic potential of colon cancer cells in vitro and in vivo. This study aims to explore the mechanisms that transgelin participates in the metastasis of colon cancer cells. -

Greg's Awesome Thesis
Analysis of alignment error and sitewise constraint in mammalian comparative genomics Gregory Jordan European Bioinformatics Institute University of Cambridge A dissertation submitted for the degree of Doctor of Philosophy November 30, 2011 To my parents, who kept us thinking and playing This dissertation is the result of my own work and includes nothing which is the out- come of work done in collaboration except where specifically indicated in the text and acknowledgements. This dissertation is not substantially the same as any I have submitted for a degree, diploma or other qualification at any other university, and no part has already been, or is currently being submitted for any degree, diploma or other qualification. This dissertation does not exceed the specified length limit of 60,000 words as defined by the Biology Degree Committee. November 30, 2011 Gregory Jordan ii Analysis of alignment error and sitewise constraint in mammalian comparative genomics Summary Gregory Jordan November 30, 2011 Darwin College Insight into the evolution of protein-coding genes can be gained from the use of phylogenetic codon models. Recently sequenced mammalian genomes and powerful analysis methods developed over the past decade provide the potential to globally measure the impact of natural selection on pro- tein sequences at a fine scale. The detection of positive selection in particular is of great interest, with relevance to the study of host-parasite conflicts, immune system evolution and adaptive dif- ferences between species. This thesis examines the performance of methods for detecting positive selection first with a series of simulation experiments, and then with two empirical studies in mammals and primates. -

Differential Expression of Drosophila Transgelins Throughout Development
fcell-09-648568 July 6, 2021 Time: 18:30 # 1 ORIGINAL RESEARCH published: 12 July 2021 doi: 10.3389/fcell.2021.648568 Differential Expression of Drosophila Transgelins Throughout Development Katerina M. Vakaloglou1†, Maria Mouratidou1†, Athina Keramidioti1,2 and Christos G. Zervas1* 1 Center of Basic Research, Biomedical Research Foundation, Academy of Athens, Athens, Greece, 2 Department of Biochemistry and Biotechnology, University of Thessaly, Larissa, Greece Transgelins are a conserved family of actin-binding proteins involved in cytoskeletal remodeling, cell contractility, and cell shape. In both mammals and Drosophila, three genes encode transgelin proteins. Transgelins exhibit a broad and overlapping expression pattern, which has obscured the precise identification of their role in development. Here, we report the first systematic developmental analysis of all Drosophila transgelin proteins, namely, Mp20, CG5023, and Chd64 in the Edited by: living organism. Drosophila transgelins display overall higher sequence identity with Lei-Miao Yin, mammalian TAGLN-3 and TAGLN-2 than with TAGLN. Detailed examination in different Shanghai University of Traditional Chinese Medicine, China developmental stages revealed that Mp20 and CG5023 are predominantly expressed in Reviewed by: mesodermal tissues with the onset of myogenesis and accumulate in the cytoplasm Cedric Soler, of all somatic muscles and heart in the late embryo. Notably, at postembryonic Clermont Université, France Simone Diestel, developmental stages, Mp20 and CG5023 are detected in the gut’s circumferential University of Bonn, Germany muscles with distinct subcellular localization: Z-lines for Mp20 and sarcomere and Rajesh Gunage, nucleus for CG5023. Only CG5023 is strongly detected in the adult fly in the abdominal, Boston Children’s Hospital, United States leg, and synchronous thoracic muscles. -

KIF23 Enhances Cell Proliferation in Pancreatic Ductal Adenocarcinoma and Is a Potent Therapeutic Target
1394 Original Article Page 1 of 15 KIF23 enhances cell proliferation in pancreatic ductal adenocarcinoma and is a potent therapeutic target Chun-Tao Gao1#, Jin Ren2#, Jie Yu1,3#, Sheng-Nan Li1, Xiao-Fan Guo1, Yi-Zhang Zhou1 1Department of Pancreatic Cancer, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Tianjin Key Laboratory of Cancer Prevention and Therapy, Tianjin’s Clinical Research Center for Cancer, Tianjin, China; 2Shanxi Bethune Hospital, Shanxi Academy of Medical Sciences, Taiyuan, China; 3The First Hospital of Shanxi Medical University, Taiyuan, China Contributions: (I) Conception and design: CT Gao, J Ren; (II) Administrative support: CT Gao; (III) Provision of study materials or patients: J Yu; (IV) Collection and assembly of data: J Ren, SN Li, YZ Zhou; (V) Data analysis and interpretation: XF Guo, J Ren; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. #These authors contributed equally to this work. Correspondence to: Chun-Tao Gao. Department of Pancreatic Cancer, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Tianjin Key Laboratory of Cancer Prevention and Therapy, Tianjin’s Clinical Research Center for Cancer, Huan-hu-xi Road, He-xi District, Tianjin 300060, China. Email: [email protected]. Background: In recent research, high expression of kinesin family member 23 (KIF23), one of the kinesin motor proteins involved in the regulation of cytokinesis, has been shown to be related to poor prognosis in glioma and paclitaxel-resistant gastric cancer, as a results of the enhancement of proliferation, migration, and invasion. In this study, we analyzed the role of KIF23 in the progression of pancreatic ductal adenocarcinoma. -

Genetic Mutations and Molecular Mechanisms of Fuchs Endothelial
Liu et al. Eye and Vision (2021) 8:24 https://doi.org/10.1186/s40662-021-00246-2 REVIEW Open Access Genetic mutations and molecular mechanisms of Fuchs endothelial corneal dystrophy Xuerui Liu1†, Tao Zheng1†, Chuchu Zhao1, Yi Zhang1, Hanruo Liu2, Liyuan Wang1* and Ping Liu1* Abstract Background: Fuchs endothelial corneal dystrophy is a hereditary disease and the most frequent cause of corneal transplantation in the worldwide. Its main clinical signs are an accelerated decrease in the number of endothelial cells, thickening of Descemet’s membrane and formation of guttae in the extracellular matrix. The cornea’s ability to maintain stromal dehydration is impaired, causing painful epithelial bullae and loss of vision at the point when the amount of corneal endothelial cells cannot be compensated. At present, apart from corneal transplantation, there is no other effective treatment that prevents blindness. Main text: In this review, we first summarized the mutations of COL8A2, TCF4, TCF8, SLC4A11 and AGBL1 genes in Fuchs endothelial corneal dystrophy. The molecular mechanisms associated with Fuchs endothelial corneal dystrophy, such as endoplasmic reticulum stress and unfolded protein response pathway, oxidative stress, mitochondrial dysregulation pathway, apoptosis pathway, mitophagy, epithelial-mesenchymal transition pathway, RNA toxicity and repeat-associated non-ATG translation, and other pathogenesis, were then explored. Finally, we discussed several potential treatments related to the pathogenesis of Fuchs endothelial corneal dystrophy, which may be the focus of future research. Conclusions: The pathogenesis of Fuchs endothelial corneal dystrophy is very complicated. Currently, corneal transplantation is an important method in the treatment of Fuchs endothelial corneal dystrophy. It is necessary to continuously explore the pathogenesis of Fuchs endothelial corneal dystrophy and establish the scientific foundations for the development of next-generation corneal therapeutics. -

Polyclonal Antibody to Transgelin (TAGLN) (C-Term) - Aff - Purified
OriGene Technologies, Inc. OriGene Technologies GmbH 9620 Medical Center Drive, Ste 200 Schillerstr. 5 Rockville, MD 20850 32052 Herford UNITED STATES GERMANY Phone: +1-888-267-4436 Phone: +49-5221-34606-0 Fax: +1-301-340-8606 Fax: +49-5221-34606-11 [email protected] [email protected] AP16212PU-N Polyclonal Antibody to Transgelin (TAGLN) (C-term) - Aff - Purified Alternate names: 22 kDa actin-binding protein, SM22, Smooth muscle protein 22-alpha, WS3-10 Quantity: 0.1 mg Concentration: 0.5 mg/ml Background: TAGLN is a transformation and shape-change sensitive actin cross-linking/gelling protein found in fibroblasts and smooth muscle. Its expression is down-regulated in many cell lines, and this down-regulation may be an early and sensitive marker for the onset of transformation. A functional role of this protein is unclear. Uniprot ID: Q01995 NCBI: NP_003177.2 GeneID: 6876 Host: Goat Immunogen: Peptide with sequence from the C-Terminus of the protein sequence according to NP_003177.2. Genename: TAGLN AA Sequence: C-MTGYGRPRQIIS Remarks: NP_003177.2 and NP_001001522.1 represent identical protein. Format: State: Liquid purified Ig fraction Purification: Ammonium Sulphate Precipitation followed by Antigen Affinity Chromatography using the immunizing peptide Buffer System: Tris saline, pH~7.3 Preservatives: 0.02% Sodium Azide Stabilizers: 0.5% BSA Applications: Peptide ELISA: 1/8000 (Detection Limit). Western blot: 0.01-0.03 mg/ml. Approx 24kDa band observed in Human Placenta lysates (calculated MW of 22.6kDa according to NP_003177.2). Other applications not tested. Optimal dilutions are dependent on conditions and should be determined by the user. -

Transcriptomic Landscape of Early Age Onset of Colorectal Cancer Identifies
www.nature.com/scientificreports OPEN Transcriptomic landscape of early age onset of colorectal cancer identifes novel genes and pathways in Indian CRC patients Manish Pratap Singh, Sandhya Rai, Nand K. Singh & Sameer Srivastava* Past decades of the current millennium have witnessed an unprecedented rise in Early age Onset of Colo Rectal Cancer (EOCRC) cases in India as well as across the globe. Unfortunately, EOCRCs are diagnosed at a more advanced stage of cancer. Moreover, the aetiology of EOCRC is not fully explored and still remains obscure. This study is aimed towards the identifcation of genes and pathways implicated in the EOCRC. In the present study, we performed high throughput RNA sequencing of colorectal tumor tissues for four EOCRC (median age 43.5 years) samples with adjacent mucosa and performed subsequent bioinformatics analysis to identify novel deregulated pathways and genes. Our integrated analysis identifes 17 hub genes (INSR, TNS1, IL1RAP, CD22, FCRLA, CXCL3, HGF, MS4A1, CD79B, CXCR2, IL1A, PTPN11, IRS1, IL1B, MET, TCL1A, and IL1R1). Pathway analysis of identifed genes revealed that they were involved in the MAPK signaling pathway, hematopoietic cell lineage, cytokine–cytokine receptor pathway and PI3K-Akt signaling pathway. Survival and stage plot analysis identifed four genes CXCL3, IL1B, MET and TNS1 genes (p = 0.015, 0.038, 0.049 and 0.011 respectively), signifcantly associated with overall survival. Further, diferential expression of TNS1 and MET were confrmed on the validation cohort of the 5 EOCRCs (median age < 50 years and sporadic origin). This is the frst approach to fnd early age onset biomarkers in Indian CRC patients. -

Monoclonal Antibody to Transgelin (SM22-Alpha)(Clone : TAGLN/247)
9853 Pacific Heights Blvd. Suite D. San Diego, CA 92121, USA Tel: 858-263-4982 Email: [email protected] 36-1708: Monoclonal Antibody to Transgelin (SM22-alpha)(Clone : TAGLN/247) Clonality : Monoclonal Clone Name : TAGLN/247 Application : FACS,IF,IHC Reactivity : Human, Mouse Gene : TAGLN Gene ID : 6876 Uniprot ID : Q01995 Format : Purified Alternative Name : TAGLN,SM22,WS3-10 Isotype : Mouse IgG1 Kappa Immunogen Information : Recombinant full-length human transgelin (TAGLN) protein. Description This MAb recognizes a 22kDa protein, identified as Transgelin, also designated SM22-alpha. It may cross-react with SM22- beta. Transgelin is expressed abundantly in smooth muscle cells. The human transgelin gene encodes a 201 amino acid protein that contains nuclear factor-binding motifs known to regulate transcription in smooth muscle. During embryogenesis, transgelin is expressed in smooth, cardiac and skeletal muscle, but is restricted during late fetal development and adulthood to all vascular and visceral smooth muscle cells and low levels of expression in heart. Transgelin is down regulated in several transformed cell lines, indicating that a reduction of transgelin expression may be an early indicator of the onset of transformation. Transgelin also binds Actin, causing Actin fibers to gel within minutes of binding. Binding of transgelin to Actin occurs at a ratio of 1:6 Actin monomers. Product Info Amount : 100 µg Purification : Affinity Chromatography 100 µg in 500 µl PBS containing 0.05% BSA and 0.05% sodium azide. Sodium azide is highly Content : toxic. Store the antibody at 4°C; stable for 6 months. For long-term storage; store at -20°C. Avoid Storage condition : repeated freeze and thaw cycles. -

Transgelin Increases Metastatic Potential of Colorectal Cancer Cells
Zhou et al. BMC Cancer (2016) 16:55 DOI 10.1186/s12885-016-2105-8 RESEARCH ARTICLE Open Access Transgelin increases metastatic potential of colorectal cancer cells in vivo and alters expression of genes involved in cell motility Hui-min Zhou1,2,3, Yuan-yuan Fang1,2, Paul M. Weinberger4,5, Ling-ling Ding6, John K. Cowell5, Farlyn Z. Hudson7, Mingqiang Ren5, Jeffrey R. Lee8, Qi-kui Chen2, Hong Su2, William S. Dynan7,9* and Ying Lin1,2* Abstract Background: Transgelin is an actin-binding protein that promotes motility in normal cells. Although the role of transgelin in cancer is controversial, a number of studies have shown that elevated levels correlate with aggressive tumor behavior, advanced stage, and poor prognosis. Here we sought to determine the role of transgelin more directly by determining whether experimental manipulation of transgelin levels in colorectal cancer (CRC) cells led to changes in metastatic potential in vivo. Methods: Isogenic CRC cell lines that differ in transgelin expression were characterized using in vitro assays of growth and invasiveness and a mouse tail vein assay of experimental metastasis. Downstream effects of transgelin overexpression were investigated by gene expression profiling and quantitative PCR. Results: Stable overexpression of transgelin in RKO cells, which have low endogenous levels, led to increased invasiveness, growth at low density, and growth in soft agar. Overexpression also led to an increase in the number and size of lung metastases in the mouse tail vein injection model. Similarly, attenuation of transgelin expression in HCT116 cells, which have high endogenous levels, decreased metastases in the same model. -
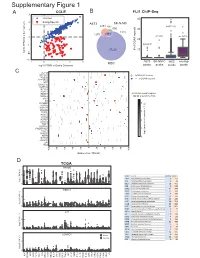
Supplementary Figure 1
6XSSOHPHQWDU\)LJXUH CCLE B FLI1 ChIP-Seq A 3 FRPPRQ 40 (ZLQJ6SHFLILF 2 A673 6.10& 477 Q 690 30 0 UHSHDWV Q Q A 20 í Q 57226 RI**$ ORJ)3.0LQRWKHUFDQFHUV í í A673 6.10& 06& RYHUODS í 0 23 06& ORJ)3.0LQ(ZLQJ6DUFRPD SHDNV SHDNV SHDNV SHDNV & ./) 67($3 (:6)/,ERXQG 3'(% ! **$AUHSHDWV 3335$ ABI3 '1$-& '863 13<5 .&1( 5%0 DOO**$AUHSHDWUHJLRQV 1.; NEDURXQGWKH766 9$9 ;* .&1$% 4 .&1$ 51) 0<20 3 355/ 355T4 &'$ $'5% 2 '&'& 711, 8*7$ 3+263+2 )(=) *1*7 ORJ QXPEHURIUHSHDWV A571 0 13<5 /2;+' /,3, 5$; 0 40 60 20 GLVWDQFHIURP766 NE D TCGA 35$0( 5 code cancer normal tumor LAML Acute Myeloid Leukemia 0 151 ORJ )3.0 ACC AdrenocoƌƟcal carcinoma 0 79 0 BLCA Bladder Urothelial Carcinoma 19 411 LGG Brain Lower Grade Glioma 0 529 COAD Colon adenocarcinoma 41 464 5%0 ESCA Esophageal carcinoma 11 162 GBM Glioblastoma muůƟforme 5 168 5 KICH Kidney Chromophobe 24 65 KIRP Kidney renal papillary cell carcinoma 32 289 LIHC Liver hepatocellular carcinoma 50 374 ORJ )3.0 LUAD Lung adenocarcinoma 59 526 0 LUSC Lung squamous cell carcinoma 49 501 DLBC Lymphoid Neoplasm Diīuse Large B-cell Lym 042 /,3, MESO Mesothelioma 0 1 OV Ovarian serous cystadenocarcinoma 0 379 PAAD PancreĂƟc adenocarcinoma 4 178 5 PCPG Pheochromocytoma and Paraganglioma 3 150 PRAD Prostate adenocarcinoma 52 499 READ Rectum adenocarcinoma 1 6 ORJ )3.0 SARC Sarcoma 0 35 0 SKCM Skin Cutaneous Melanoma 1 471 STAD Stomach adenocarcinoma 32 375 /2;+' WXPRU TGCT dĞƐƟĐƵůĂƌ'ĞƌŵĞůůdƵŵŽƌƐ 0156 THCA Thyroid carcinoma 58 510 QRUPDO UCS Uterine Carcinosarcoma 0 56 5 ORJ )3.0 0 / / 29 $&& 8&6 /** *%0