A Case Study in the Fern Myriopteris Gracilis (Pteridaceae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Appendix E High-Potential Historic Sites
APPENDIX E HIGH-POTENTIAL HISTORIC SITES National Trails System Act, SEC. 12. [16USC1251] As used in this Act: (1) The term “high-potential historic sites” means those historic sites related to the route, or sites in close proximity thereto, which provide opportunity to interpret the historic significance of the trail during the period of its major use. Criteria for consideration as high-potential sites include historic sig nificance, presence of visible historic remnants, scenic quality, and relative freedom from intrusion.. Mission Ysleta, Mission Trail Indian and Spanish architecture including El Paso, Texas carved ceiling beams called “vigas” and bell NATIONAL REGISTER tower. Era: 17th, 18th, and 19th Century Mission Ysleta was first erected in 1692. San Elizario, Mission Trail Through a series of flooding and fire, the mis El Paso, Texas sion has been rebuilt three times. Named for the NATIONAL REGISTER patron saint of the Tiguas, the mission was first Era: 17th, 18th, and 19th Century known as San Antonio de la Ysleta. The beauti ful silver bell tower was added in the 1880s. San Elizario was built first as a military pre sidio to protect the citizens of the river settle The missions of El Paso have a tremendous ments from Apache attacks in 1789. The struc history spanning three centuries. They are con ture as it stands today has interior pillars, sidered the longest, continuously occupied reli detailed in gilt, and an extraordinary painted tin gious structures within the United States and as ceiling. far as we know, the churches have never missed one day of services. -

The Lower Permian Abo Formation in the Fra Cristobal and Caballo Mountains, Sierra County, New Mexico Spencer G
New Mexico Geological Society Downloaded from: http://nmgs.nmt.edu/publications/guidebooks/63 The Lower Permian Abo Formation in the Fra Cristobal and Caballo Mountains, Sierra County, New Mexico Spencer G. Lucas, Karl Krainer, Dan S. Chaney, William A. DiMichele, Sebastian Voigt, David S. Berman, and Amy C. Henrici, 2012, pp. 345-376 in: Geology of the Warm Springs Region, Lucas, Spencer G.; McLemore, Virginia T.; Lueth, Virgil W.; Spielmann, Justin A.; Krainer, Karl, New Mexico Geological Society 63rd Annual Fall Field Conference Guidebook, 580 p. This is one of many related papers that were included in the 2012 NMGS Fall Field Conference Guidebook. Annual NMGS Fall Field Conference Guidebooks Every fall since 1950, the New Mexico Geological Society (NMGS) has held an annual Fall Field Conference that explores some region of New Mexico (or surrounding states). Always well attended, these conferences provide a guidebook to participants. Besides detailed road logs, the guidebooks contain many well written, edited, and peer-reviewed geoscience papers. These books have set the national standard for geologic guidebooks and are an essential geologic reference for anyone working in or around New Mexico. Free Downloads NMGS has decided to make peer-reviewed papers from our Fall Field Conference guidebooks available for free download. Non-members will have access to guidebook papers two years after publication. Members have access to all papers. This is in keeping with our mission of promoting interest, research, and cooperation regarding geology in New Mexico. However, guidebook sales represent a significant proportion of our operating budget. Therefore, only research papers are available for download. -

Overview for Geologic Field-Trip Guides to Mount Mazama, Crater Lake Caldera, and Newberry Volcano, Oregon
Overview for Geologic Field-Trip Guides to Mount Mazama, Crater Lake Caldera, and Newberry Volcano, Oregon Scientific Investigations Report 2017–5022–J U.S. Department of the Interior U.S. Geological Survey Cover (top photo): View east-northeast from Garfield Peak on the south rim of Crater Lake caldera. Peak on skyline is 8,929 feet (2,722 meters) Mount Scott, an ~420 thousand years before present (ka) dacite stratovolcano considered to be part of Mount Mazama, the volcano that collapsed during the caldera-forming eruption ~7,700 years ago. The caldera walls in this view expose Mazama lava flows and fragmental deposits from as old as ~400 ka at Phantom Cone, adjacent to tiny Phantom Ship island, to as young as ~27 ka at Redcloud Cliff, the V-shaped face at the top of the wall left of center. The beheaded glacial valley of Kerr Notch, the low point on the caldera rim, is seen between Phantom Ship and Mount Scott. Photograph by Carly McLanahan. Cover (bottom photo): Newberry Volcano, Oregon, is the largest volcano in the Cascades volcanic arc. This north-facing view taken from the volcano’s peak, Paulina Peak (elevation 7,984 feet), encompasses much of the volcano’s 4-by-5-milewide central caldera, a volcanic depression formed in a powerful explosive eruption about 75,000 years ago. The caldera’s two lakes, Paulina Lake (left) and the slightly higher East Lake (right), are fed in part by active hot springs heated by molten rock (magma) deep beneath the caldera. The Central Pumice Cone sits between the lakes. -
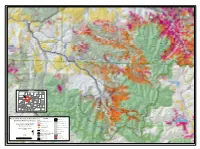
Forest Wide Hazardous Tree Removal and Fuels Reduction Project
107°0'0"W VAIL k GYPSUM B e 6 u 6 N 1 k 2 k 1 h 2 e . e 6 . .1 I- 1 o 8 70 e c f 7 . r 0 e 2 2 §¨¦ e l 1 0 f 2 u 1 0 3 2 N 4 r r 0 1 e VailVail . 3 W . 8 . 1 85 3 Edwards 70 1 C 1 a C 1 .1 C 8 2 h N 1 G 7 . 7 0 m y 1 k r 8 §¨¦ l 2 m 1 e c . .E 9 . 6 z W A T m k 1 5 u C 0 .1 u 5 z i 6. e s 0 C i 1 B a -7 k s 3 2 .3 e e r I ee o C r a 1 F G Carterville h r e 9. 1 6 r g 1 N 9 g 8 r e 8 r y P e G o e u l Avon n C 9 N C r e n 5 ch w i r 8 .k2 0 N n D k 1 n 70 a tt e 9 6 6 8 G . c 7 o h 18 1 §¨¦ r I-7 o ra West Vail .1 1 y 4 u h 0 1 0. n lc 7 l D .W N T 7 39 . 71 . 1 a u 1 ch W C k 0 C d . 2 e . r e 1 e 1 C st G e e . r 7 A Red Hill R 3 9 k n s e 5 6 7 a t 2 . -

Chiricahua National Monument Historic Designed Landscape Historic Name
NPS Form 10-900 OMB No. 1024-0018 (Oct. 1990) United States Department of the Interior National Park Service National Register of Historic Places Registration Form This form is for use in nominating or requesting determinations for individual properties and districts. See instructions in How to Complete the National Register of Historic Places Registration Form (National Register Bulletin 16A). Complete each item by marking "x" in the appropriate box or by entering the information requested. If an item does not apply to the property being nominated, enter "N/A" for "not applicable." For functions, architectural classification, materials, and areas of significance, enter only categories and subcategories from the instructions. Place additional entries and narrative items on continuation sheets (NPS Form 10-900a). Use a typewriter, word processor, or computer, to complete all items. 1. Name of Property Chiricahua National Monument Historic Designed Landscape historic name other name/site number Wonderland of Rocks; Rhyolite Park; The Pinnacles; Say Yahdesut “Point of Rocks” 2. Location street & number: Chiricahua National Monument (CHIR) 12856 E. Rhyolite Canyon Road _____not for publication city/town: Willcox___________________________________________________________ _X_ vicinity state: Arizona_____ code: AZ __________ county: Cochise_________ code: 003_____ zip code: 85643___ 3. State/Federal Agency Certification As the designated authority under the National Historic Preservation Act, as amended, I hereby certify that this ¨ nomination ¨ request for determination of eligibility meets the documentation standards for registering properties in the National Register of Historic Places an meets the procedural and professional requirements set forth in 36 CFR Part 60. In my opinion, the property ¨ meets ¨ does not meet the National Register criteria. -

Salida Buena Vista
CHAFFEE COUNTY, COLORADO R 82 W R 81 W R 80 W R 79 W R 78 W R 77 W R 76 W South Peak Mt Elbert Casco Peak Black Mountain Bull Hill Independence Mountain Parry Peak T 11 S Lower Lake Upper Lake Monitor Rock T 11 S Star Mountain LAKE COUNTY Ouray Peak l Y Grizzly Lake 32 33 34 35 36 31 32 33 36 31 32 33 34 35 33 34 T 34 32 Grizzly Peak 35 36 31 35 l 31 G r a n i t e 398-D G r a n i t e 2905 l Twin Peaks +$ N 2903 l Rinker Peak 2901 4 2899 6 5 3 2 1 6 5 6 4 3 1 Garfield Peak 4 3 2 1 5 2 6 4 2 l La Plata Peak 5 3 U l lRed Mountain 7 8 O 7 9 10 11 12 9 Willis Lake l 8 9 10 11 8 10 Mt H11ope 12 7 12 Crystal Lake lQuail Mountain 7 8 9 10 11 Middle Mountain Clear Creek Reservoir l C 3 18 17 16 15 13 16 14 18 17 15 14 18 17 16 15 14 13 13 18 15 14 0 $+390 17 16 13 N 5 V i c k s b u r g 7 I V i c k s b u r g 9 T 12 S 21 0 20 19 20 19 21 022 23 24 $+388 23 24 19 20 21 022 23 22 0 24 19 20 21 22 23 24 19 20 l T 12 S K 3 0 West Buffalo Peak 0 ¤£204 21 l $+371 P Antero Reservoir W i n f i e l d East Buffalo Peak T W3i n f i e l d 30 Winfield Pea2k9 28 27 26 I 3 A l 25 3 25 30 29 26 28 27 26 27 26 l 25 30 29 28 25 30 29 28 l Cross Mountain Waverly Mountain 1 R P Jenkins Mountain Middle Mountain 371 K l $+ Virginia Peak 1 l Mt Oxford l 36 31 32 33 34 35 36 31 32 35 Browns Peak 33 34 35 36 C 32 34 35 Mt Belford 31 33 36 31 032 33 l Waupaca Reservoir O l 3 2 U Rainbow Lake 1 6 5 4 3 2 1 l6Missouri Mountain5 4 2 Huron Peak Cloyses Lake 3 1 6 5 4 3 2 1 6 N l 5 4 Lois Lake Granite Mountain T l Iowa Peak l l 1 6 386 Y 12 $+ Marmot Peak 9 7 8 9 10 11 12 7 8 -

Silver and Gold Occurrences in New Mexico by Robert M
New Mexico Bureau of Mines & Mineral Resources Resource Map 15 March 1986 Silver and gold occurrences in New Mexico by Robert M. North and Virginia T. Mclemore New Mexico Bureau of Mines and Mineral Resources, Socorro, New Mexico 87801 Abstract Anomalous amounts of silver and/or gold have been identified in 153 mining districts or prospect areas of New Mexico. Production from most of these occurrences has been small; figures or estimates are given when known. Thirty-five districts have produced in excess of 10,000 troy ounces of gold or 200,000 ounces of silver. Silver and gold in New Mexico occur in 14 distinct types of deposits ranging in age from Precam brian (Proterozoic) to Recent. Mid-Tertiary to Recent deposits include the placer, volcanic-epithermal, supergene copper-uranium (silver), and Great Plains Margin types. Late Cretaceous to mid-Tertiary deposits include the sedimentary-hydrothermal barite-fluorite-galena, carbonate-hosted silver (lead manganese ), lead-zinc and copper skarn, Laramide vein, and prophyry-copper types. Late Paleozoic to early Mesozoic deposits include the sedimentary-copper type and possibly some of the Permian Mississippi Valley type. Precambrian deposits include the vein and replacement and Precambrian massive-sulfide types. Deposits that have produced significant silver and/or gold as the primary product are the placer, volcanic-epithermal, Great Plains Margin, carbonate-hosted silver, and Lar amide vein types. Deposits that have produced significant precious metals as a byproduct of base metal mining include the carbonate-hosted lead-zinc, copper skarn, and porphyry-copper types. Introduction Precious metals in varying degrees of importance or 14 ppm (0.41 oz/ton) silver. -

Crater Lake National Park Oregon
DEPARTMENT OF THE INTERIOR HUBERT WORK. SECRETARY NATIONAL PARK SERVICE STEPHEN T. MATHER. DIRECTOR RULES AND REGULATIONS CRATER LAKE NATIONAL PARK OREGON PALISADE POINT, MOUNT SCOTT IN THE DISTANCE 1923 Season from July 1 to September 30 THE PHANTOM SHIP. FISHING IS EXCELLENT IN CRATER LAKE. THE NATIONAL PARKS AT A GLANCE. [Number, 19; total area, 11,372 square miles.] Area in National parks in Distinctive characteristics. order of creation. Location. squaro miles. Hot Springs Middle Arkansas li 40 hot springs possessing curative properties- 1832 Many hotels and boarding houses—20 bath houses under public control. Yellowstone Northwestern Wyo 3.348 More geysers than in all rest of world together- 1872 ming. Boiling springs—Mud volcanoes—Petrified for ests—Grand Canyon of the Yellowstone, remark able for gorgeous coloring—Large lakes—Many largo streams and waterfalls—Vast wilderness, greatest wild bird and animal preserve in world— Exceptional trout fishing. Sequoia. Middle eastern Cali 252 The Big Tree National Park—several hundred 1S90 fornia. sequoia trees over 10 feet in diameter, some 25 to 36 feet, hi diameter—Towering mountain ranges- Startling precipices—Mile long cave of delicate beauty." Yosemito Middle eastern Cali 1,125 Valley of world-famed beauty—Lofty chits—Ro 1890 fornia. mantic vistas—Many waterfalls of extraordinary height—3 groves of big trees—High Sierra— Waterwhcol falls—Good trout fishing. General Grant Middle eastern Cali 4 Created to preserve the celebrated General Grant 1S90 fornia. Tree, 3* feet in diameter—6 miles from Sequoia National Park. Mount Rainier ... West central Wash 321 Largest accessible single peak glacier system—28 1899 ington. -

Geologic Map of the Truth Or Consequences 30' X 60' Minute
1 GeoloPic MaD of the Truth or Conseauences 30 x 60 Minute Ouadrande (1:lOO.OOO scale) Richard W. Harrison, Richard P. Lozinsky, Ted L. Eggleston, and William C. McIntosh INTRODUCTION The geologic map of the Truth or Consequences 30 x 60 minute quadrangle has grown out of the Ph.D. dissertations and related work of the authors in south-central New Mexico. Funding and support for this work was provided by New Mexico Bureau of Mines and Mineral Resources. And while the authors have mapped extensive portionsof the sheet in either detailed or reconnaissance fashion, large areas of the sheet have been compiled from existing maps (see attached diagram for sources). This is an in-progress status report. Many aspects of the complex geology found in this area are yet to be worked out. Descriptions of stratigraphy and swcture provided in this report are brief. For more details, reference is given to Eggleston (1986), McIntosh (1989), Harrison (1990), and Lozinsky and Hawley (1986). STRATIGRAPHY QC Quaternarycolluvium QcaQuaternary alluvium andcolluvium, undivided QPgQuaternary piedmont slopedeposits To basalts:Tertiary Pliocene, approximately 4.2-4.8 Ma (IUAr dates) in Winston-Las Animas grabens, approximately2.1 Ma (K/Ardate) in Cutter Sag area, the younger group upon a pediment surface in the Cutter Sag area. Tsf Santa Fe Group: Oligocene and younger basin fill of the Rio Grande drainage west of the Continental Divide: basal beds are intercalated with Oligocene volcanic rocks,are moderately well indurated, more voluminous, younger beds overlie basal beds with angular unconformity andare generally near horizontalin attitude. The Santa Fe Group is dominantly volcaniclastic and heterolithic. -

Bulletin 39: the Metal Resources of New Mexico and Their Economic
BULLETIN 3 9 The Metal Resources of New Mexico and Their Economic Features Through 1954 A revision of Bulletin 7, by Lasky and Wootton, with detailed information for the years 1932-1954 BY EUGENE CARTER ANDERSON 1957 STATE BUREAU OF MINES AND MINERAL RESOURCES NEW MEXICO INSTITUTE OF MINING & TECHNOLOGY CAMPUS STATION SOCORRO, NEW MEXICO NEW MEXICO INSTITUTE OF MINING & TECHNOLOGY E. J. Workman, President STATE BUREAU OF MINES AND MINERAL RESOURCES Alvin J. Thompson, Director THE REGENTS MEMBERS EX OFFICIO THE HONORABLE EDWIN L. MECHEM………...Governor of New Mexico MRS. GEORGIA L. LUSK ......................Superintendent of Public Instruction APPOINTED MEMBERS ROBERT W. BOTTS ....................................................................Albuquerque HOLM O. BURSUM, JR. .....................................................................Socorro THOMAS M. CRAMER .................................................................... Carlsbad JOHN N. MATHEWS, JR. ...................................................................Socorro RICHARD A. MATUSZESKI ......................................................Albuquerque Contents Page INTRODUCTION .......................................................................................................... 1 Purpose and Scope of Bulletin ..................................................................................... 1 Other Reports Dealing With the Geology and Mineral Resources of New Mexico ...................................................................................................... -

Hike the Parks
Hike the Parks ~Rulebook~ Hello! Welcome to Hike the Parks, a U.S. National Parks game with authentic hikes and cool facts. Roll the dice to visit new states, territories and Park Sites; learn cool facts at the Visitor’s Center; go on hikes; and take pictures! Brief Summary: 1. The winner is the first player to reach 200 Joy. Change the amount to shorten or lengthen the game. 2. Joy is earned by going on hikes, taking pictures while on hikes, meeting nice hitchhikers and optionally earning badges along the way. 3. Players roll two die to travel to different U.S. states. For example, if a 2 & 3 is rolled, a player can move across a maximum of 5 state borders to visit a park for that turn. If a player only crosses 3 of the 5 allowed borders for that turn, the remaining 2 CANNOT be carried over to another turn. 4. A player gains one Energy and one Water for each state border crossed on their turn. For example, traveling from Washington to Alaska gains two Energy and two Water. 5. After crossing at least one border, a player must draw one On the Road card for that turn. If the player remains in the same state, they do not need to pick up an On the Road card. 6. After visiting a new state or deciding to stay in the same state, a player may visit one Park Site (or stay in the same Park Site) and “visit its Visitors Center” by drawing a Visitor’s Center Trivia card to learn about a random National Park Site. -

Metamorphism
Contents AΒSTRACT 7 STRUCTURAL GEOLOGY 31 INTRODUCTION 7 SMP BLOCK 31 METAMORPHIC ROCKS 11 SAN ANDRES MOUNTAINS 32 MICA-QUARTZ SCHIST AND PHYLLITE 11 PEDERNAL HILLS 32 QUARTZITE AND RELATED ROCKS 13 LADRON MOUNTAINS 33 MAFIC META-IGNEOUS ROCKS 16 OTHER AREAS 33 SILICEOUS META-IGNEOUS ROCKS 18 CONCLUSIONS 33 GNEISS 20 METAMORPHISM 34 STRATIGRAPHY 21 GEOCHRONOLOGY 37 GRANITIC ROCKS 24 GEOCHEMISTRY 38 NORTH AND SOUTH SANDIA PLUTONS 25 MAFIC META-IGNEOUS ROCKS 38 MANZANITA PLUTON 26 SILICEOUS META-IGNEOUS ROCKS AND GRANITIC ROCKS 40 ΟJΙΤΑ AND MONTE LARGO PLUTONS 26 High-Ca group 40 PRIEST PLUTON 27 High-K group 42 LOS PINOS PLUTON 27 High-Si group 43 SEPULTURA PLUTON 27 Effects of alteration 44 PEDERNAL PLUTON 27 FELDSPATHIC QUARTZITES AND ARKOSITES 45 LADRON PLUTON 28 MAGMA ORIGIN 47 CAPIROTE PLUTON 28 TECTONIC SETTING AND LA JOYITA PLUTON 28 GEOLOGIC EVOLUTION 48 POLVADERA PLUTON 28 MINERAL DEPOSITS 50 MAGDALENA PLUTON 28 REFERENCES 51 OSCURA AND CAPITOL PEAK PLUTONS 28 APPENDICES 54 MOCKINGBIRD GAP PLUTON 29 1—Composite stratigraphic sections 54 STRAWBERRY PEAK AND MAYBERRY PLUTONS 30 2—Major-element chemical analyses (microfiche SAN ANDRES PLUTON 30 in pocket) MINERAL HILL PLUTON 30 3—Trace-element contents (microfiche in pocket) GRANITIC ROCKS OF THE CABALLO MOUNTAINS AND 4—Sample locations (microfiche in pocket) RED HILLS 30 INDEX 57 MISCELLANEOUS PLUTONS 30 Tables 1—Relative percentages of metamorphic rocks in Precambrian 9—Geochemical classification of Precambrian siliceous meta- sections 11 igneous rocks and granitic plutons 40 2—Modal