Litsea Complex, Lauraceae) Inferred from Sequences of the Chloroplast Gene Matk and Nuclear Ribosomal DNA ITS Regions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

DPR Journal 2016 Corrected Final.Pmd
Bul. Dept. Pl. Res. No. 38 (A Scientific Publication) Government of Nepal Ministry of Forests and Soil Conservation Department of Plant Resources Thapathali, Kathmandu, Nepal 2016 ISSN 1995 - 8579 Bulletin of Department of Plant Resources No. 38 PLANT RESOURCES Government of Nepal Ministry of Forests and Soil Conservation Department of Plant Resources Thapathali, Kathmandu, Nepal 2016 Advisory Board Mr. Rajdev Prasad Yadav Ms. Sushma Upadhyaya Mr. Sanjeev Kumar Rai Managing Editor Sudhita Basukala Editorial Board Prof. Dr. Dharma Raj Dangol Dr. Nirmala Joshi Ms. Keshari Maiya Rajkarnikar Ms. Jyoti Joshi Bhatta Ms. Usha Tandukar Ms. Shiwani Khadgi Mr. Laxman Jha Ms. Ribita Tamrakar No. of Copies: 500 Cover Photo: Hypericum cordifolium and Bistorta milletioides (Dr. Keshab Raj Rajbhandari) Silene helleboriflora (Ganga Datt Bhatt), Potentilla makaluensis (Dr. Hiroshi Ikeda) Date of Publication: April 2016 © All rights reserved Department of Plant Resources (DPR) Thapathali, Kathmandu, Nepal Tel: 977-1-4251160, 4251161, 4268246 E-mail: [email protected] Citation: Name of the author, year of publication. Title of the paper, Bul. Dept. Pl. Res. N. 38, N. of pages, Department of Plant Resources, Kathmandu, Nepal. ISSN: 1995-8579 Published By: Mr. B.K. Khakurel Publicity and Documentation Section Dr. K.R. Bhattarai Department of Plant Resources (DPR), Kathmandu,Ms. N. Nepal. Joshi Dr. M.N. Subedi Reviewers: Dr. Anjana Singh Ms. Jyoti Joshi Bhatt Prof. Dr. Ram Prashad Chaudhary Mr. Baidhya Nath Mahato Dr. Keshab Raj Rajbhandari Ms. Rose Shrestha Dr. Bijaya Pant Dr. Krishna Kumar Shrestha Ms. Shushma Upadhyaya Dr. Bharat Babu Shrestha Dr. Mahesh Kumar Adhikari Dr. Sundar Man Shrestha Dr. -

Gori River Basin Substate BSAP
A BIODIVERSITY LOG AND STRATEGY INPUT DOCUMENT FOR THE GORI RIVER BASIN WESTERN HIMALAYA ECOREGION DISTRICT PITHORAGARH, UTTARANCHAL A SUB-STATE PROCESS UNDER THE NATIONAL BIODIVERSITY STRATEGY AND ACTION PLAN INDIA BY FOUNDATION FOR ECOLOGICAL SECURITY MUNSIARI, DISTRICT PITHORAGARH, UTTARANCHAL 2003 SUBMITTED TO THE MINISTRY OF ENVIRONMENT AND FORESTS GOVERNMENT OF INDIA NEW DELHI CONTENTS FOREWORD ............................................................................................................ 4 The authoring institution. ........................................................................................................... 4 The scope. .................................................................................................................................. 5 A DESCRIPTION OF THE AREA ............................................................................... 9 The landscape............................................................................................................................. 9 The People ............................................................................................................................... 10 THE BIODIVERSITY OF THE GORI RIVER BASIN. ................................................ 15 A brief description of the biodiversity values. ......................................................................... 15 Habitat and community representation in flora. .......................................................................... 15 Species richness and life-form -

Antihyperglycemic Activity of Compounds Isolated from Indian Medicinal Plants
Indian Journal of Experimental Biology Vol. 48, March 2010, pp. 294-298 Antihyperglycemic activity of compounds isolated from Indian medicinal plants Akanksha a, Arvind K Srivastava b & Rakesh Maurya a* aMedicinal and Process Chemistry Division, bDivision of Biochemistry, Central Drug Research Institute, CSIR, Lucknow 226 001, India Received 4 November 2009; revised 3 December 2009 Eleven antidiabetic Indian medicinal plants were investigated in streptozotocin induced diabetic rat model and provided scientific validation to prove their antihyperglycemic activity. Antidiabetic principles from five plants were isolated. All the compounds isolated were evaluated for antihyperglycemic activity in streptozotocin induced diabetic rat model and activities were compared with standard drug metformin. Some compounds were also screened in db/db mice. Two compounds (PP-1 and PP-2) inhibited significantly the activity of PTPase-1B in an in vitro system. This might be the underlying mechanism of antihyperglycemic activity of these compounds. Keywords : Antihyperglycemic activity, Normoglycemic rat model, PTPase-1B, STZ Diabetes mellitus is characterized by group of grandis, Tinospora cordifolia, Withania coagulans metabolic disorders. Deficiency or insensitivity of and Zingiber officinale, and isolated antidiabetic insulin causes glucose to accumulate in the blood, principals from F. bengalensis , P. pinnata , leading to various complications. When breakdown of P. marsupium , T. cordifolia and W. coagulans . glucose is stopped, body uses fat and protein for producing the energy. Due to this polydipsia, Materials and Methods polyuria, polyphagia, and excessive weight loss occur. Plant materials Authenticated leaves of Aegle High blood sugar harms organs and increases risk of marmelose (collected in the month of March from heart disease. Cardiovascular disease, retinopathy, Lucknow, CDRI plant code no. -

Phylogeny and Historical Biogeography of Lauraceae
PHYLOGENY Andre'S. Chanderbali,2'3Henk van der AND HISTORICAL Werff,3 and Susanne S. Renner3 BIOGEOGRAPHY OF LAURACEAE: EVIDENCE FROM THE CHLOROPLAST AND NUCLEAR GENOMES1 ABSTRACT Phylogenetic relationships among 122 species of Lauraceae representing 44 of the 55 currentlyrecognized genera are inferredfrom sequence variation in the chloroplast and nuclear genomes. The trnL-trnF,trnT-trnL, psbA-trnH, and rpll6 regions of cpDNA, and the 5' end of 26S rDNA resolved major lineages, while the ITS/5.8S region of rDNA resolved a large terminal lade. The phylogenetic estimate is used to assess morphology-based views of relationships and, with a temporal dimension added, to reconstructthe biogeographic historyof the family.Results suggest Lauraceae radiated when trans-Tethyeanmigration was relatively easy, and basal lineages are established on either Gondwanan or Laurasian terrains by the Late Cretaceous. Most genera with Gondwanan histories place in Cryptocaryeae, but a small group of South American genera, the Chlorocardium-Mezilauruls lade, represent a separate Gondwanan lineage. Caryodaphnopsis and Neocinnamomum may be the only extant representatives of the ancient Lauraceae flora docu- mented in Mid- to Late Cretaceous Laurasian strata. Remaining genera place in a terminal Perseeae-Laureae lade that radiated in Early Eocene Laurasia. Therein, non-cupulate genera associate as the Persea group, and cupuliferous genera sort to Laureae of most classifications or Cinnamomeae sensu Kostermans. Laureae are Laurasian relicts in Asia. The Persea group -

NJZ Online Vol. 2, Issue 1
Association of Anaemia with Parasitic Infection in Pregnant Women Attending Antenatal Clinic at Koshi Zonal Hospital Manju Chaudhary and Mahendra Maharjan Central Department of Zoology, Tribhuvan University, Kirtipur, Kathmandu, Nepal For correspondence: [email protected] Abstract Intestinal parasitic infections associated with anaemia during pregnancy have direct negative impact on the health of expected mother and developing baby. In order to assess the association between anaemia and parasitic infection during pregnancy, a total of 200 stool samples from pregnant women on their first consultation to antenatal service in Koshi Zonal Hospital were collected from April to August 2012. The stool samples were examined for intestinal parasites by direct smear technique, while haemoglobin level of pregnant women were collected from laboratory record of the hospital. Out of 110 anaemic pregnant women 40(36.3%) had parasitic infection, while from 90 non-anaemic pregnant women; only 18(20%) of them were infected with intestinal parasites. The association of anaemia with intestinal parasite was statistically significant (p<0.008). The prevalence of Hookworm (76.9%) was most prevalent infection followed by Ascaris lumbricoides (73.3%) in anaemic pregnant women. The mean Haemoglobin (Hb) level of pregnant women with single parasite and with multiple infection was 10.4 ± 1.80 gm/dl (mild anaemia) and 9.81 ± 0.84 gm/dl (moderate anaemia) respectively. However, the overall prevalence of the parasitic infection among pregnant women was 58(29%). A. lumbricoides (32.3%) was the most predominant followed by Hookworm (26.1%), Giardia lamblia (21.5%), Entamoeba histolytica (10.7%), Trichuris trichiura (6.15%), Strongyloides stercoralis (1.5%) and Hymenolepis nana (1.5%). -

Isolation and Identification of Cyclic Polyketides From
ISOLATION AND IDENTIFICATION OF CYCLIC POLYKETIDES FROM ENDIANDRA KINGIANA GAMBLE (LAURACEAE), AS BCL-XL/BAK AND MCL-1/BID DUAL INHIBITORS, AND APPROACHES TOWARD THE SYNTHESIS OF KINGIANINS Mohamad Nurul Azmi Mohamad Taib, Yvan Six, Marc Litaudon, Khalijah Awang To cite this version: Mohamad Nurul Azmi Mohamad Taib, Yvan Six, Marc Litaudon, Khalijah Awang. ISOLATION AND IDENTIFICATION OF CYCLIC POLYKETIDES FROM ENDIANDRA KINGIANA GAMBLE (LAURACEAE), AS BCL-XL/BAK AND MCL-1/BID DUAL INHIBITORS, AND APPROACHES TOWARD THE SYNTHESIS OF KINGIANINS . Chemical Sciences. Ecole Doctorale Polytechnique; Laboratoires de Synthase Organique (LSO), 2015. English. tel-01260359 HAL Id: tel-01260359 https://pastel.archives-ouvertes.fr/tel-01260359 Submitted on 22 Jan 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. ISOLATION AND IDENTIFICATION OF CYCLIC POLYKETIDES FROM ENDIANDRA KINGIANA GAMBLE (LAURACEAE), AS BCL-XL/BAK AND MCL-1/BID DUAL INHIBITORS, AND APPROACHES TOWARD THE SYNTHESIS OF KINGIANINS MOHAMAD NURUL AZMI BIN MOHAMAD TAIB FACULTY OF SCIENCE UNIVERSITY -

Phylogeny, Molecular Dating, and Floral Evolution of Magnoliidae (Angiospermae)
UNIVERSITÉ PARIS-SUD ÉCOLE DOCTORALE : SCIENCES DU VÉGÉTAL Laboratoire Ecologie, Systématique et Evolution DISCIPLINE : BIOLOGIE THÈSE DE DOCTORAT Soutenue le 11/04/2014 par Julien MASSONI Phylogeny, molecular dating, and floral evolution of Magnoliidae (Angiospermae) Composition du jury : Directeur de thèse : Hervé SAUQUET Maître de Conférences (Université Paris-Sud) Rapporteurs : Susanna MAGALLÓN Professeur (Universidad Nacional Autónoma de México) Thomas HAEVERMANS Maître de Conférences (Muséum national d’Histoire Naturelle) Examinateurs : Catherine DAMERVAL Directeur de Recherche (CNRS, INRA) Michel LAURIN Directeur de Recherche (CNRS, Muséum national d’Histoire Naturelle) Florian JABBOUR Maître de Conférences (Muséum national d’Histoire Naturelle) Michael PIRIE Maître de Conférences (Johannes Gutenberg Universität Mainz) Membres invités : Hervé SAUQUET Maître de Conférences (Université Paris-Sud) Remerciements Je tiens tout particulièrement à remercier mon directeur de thèse et ami Hervé Sauquet pour son encadrement, sa gentillesse, sa franchise et la confiance qu’il m’a accordée. Cette relation a immanquablement contribuée à ma progression humaine et scientifique. La pratique d’une science sans frontière est la plus belle chose qu’il m’ait apportée. Ce fut enthousiasmant, très fructueux, et au-delà de mes espérances. Ce mode de travail sera le mien pour la suite de ma carrière. Je tiens également à remercier ma copine Anne-Louise dont le soutien immense a contribué à la réalisation de ce travail. Elle a vécu avec patience et attention les moments d’enthousiasmes et de doutes. Par la même occasion, je remercie ma fille qui a eu l’heureuse idée de ne pas naître avant la fin de la rédaction de ce manuscrit. -

118 MW Nikacchu Hydropower Project
Environmental Safeguard Monitoring Report 1st Quarterly Report December 2015 BHU: Second Green Power Development Project- 118 MW Nikacchu Hydropower Project Prepared by the Tangsibji Hydro Energy Limited for the Asian Development Bank. This environmental safeguard monitoring report is a document of the borrower. The views expressed herein do not necessarily represent those of ADB's Board of Directors, Management, or staff, and may be preliminary in nature. In preparing any country program or strategy, financing any project, or by making any designation of or reference to a particular territory or geographic area in this document, the Asian Development Bank does not intend to make any judgments as to the legal or other status of any territory or area. Environmental Safeguard Monitoring Report Reporting Period August, 2015 to November, 2015} Date December, 2015 BHU: Second Green Power Development Project: (118 MW Nikacchu Hydropower Project) Prepared by the Tangsibji Hydro Energy Limited for the Asian Development Bank This environmental safeguard monitoring report is a document of the borrower and made publicly available in accordance with ADB’s Public Communications Policy 2011 and the Safeguard Policy Statement 2009. The views expressed herein do not necessarily represent those of ADB’s Board of Directors, Management, or staff. P a g e | 1 Table of Contents Executive Summary ................................................................................................................................... 3 1.0. Introduction ..................................................................................................................................... -

Chapter 1 Introduction
CHAPTER 1 INTRODUCTION Chapter 1: Introduction 1.1 General Natural products could be any biological molecule, but the term is usually reserved for secondary metabolites or small molecules produced by an organism.1 They are not strictly necessary for the survival of the organism but they could be for the basic machinery of the fundamental processes of life.2 Many phytochemicals have been used as medicines such as for cardiovascular drugs, reserpine and digitoxin which were isolated from Rauwolfia serpentine and Digitalis purpurea.3,4 The natural antimalarial drug, quinine from Cinchona species together with the synthetic drugs, chloroquine and antabrin are still widely used until today to treat malaria.5 Plants are of great importance to many local communities in Malaysia and Asia, either used as health supplements or for therapeutic purposes. Concoctions of several species in the form of jamu are consumed daily. Herbal medicine is now becoming more popular as alternative to western medicine and is now available in many forms. As a result, there has been rapid development of herbal industries utilizing locally available plants, either wild or introduced.6 Malaysian tropical forest is home to a large number of plants and considered to be one of the oldest and richest in the world. The rich Malaysian flora provides opportunities for the discovery of novel compounds, some of which could have useful bioactivities. The use of plant extracts and phytochemicals, both with known biological activities, can be of great significance in therapeutic treatments. Lauraceae plants are known to be prolific producers of many interesting alkaloids such as the rare proaporphine-tryptamine dimers; phoebegrandines A-B and (-)-phoebescortechiniine isolated from Phoebe grandis and Phoebe scortechinii, and bisbenzylisoquinoline alkaloids; oxoperakensimines A-C and 3, 4-dihydronorstephasubine isolated from Alseodaphne perakensis and Alseodaphne corneri 1 Chapter 1: Introduction respectively. -
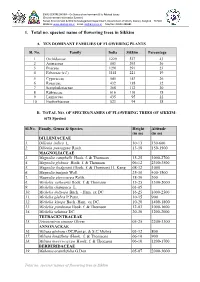
1. Total No. Species/ Name of Flowering Trees in Sikkim
ENVIS CENTRE SIKKIM – On Status of environment & Its Related Issues (Environmental Information System) Forest, Environment & Wildlife Management Department, Government of Sikkim, Deorali, Gangtok – 737102 Website: www.sikenvis.nic.in Email: [email protected] Tele/Fax: 03592-280381 1. Total no. species/ name of flowering trees in Sikkim A. TEN DOMINANT FAMILIES OF FLOWERING PLANTS Sl. No. Family India Sikkim Percentage 1 Orchidaceae 1229 527 43 2 Asteraceae 803 293 36 3 Poaceae 1291 291 23 4 Fabaceae (s.l.) 1141 221 19 5 Cyperaceae 545 143 26 6 Rosaceae 432 138 32 7 Scrophulariaceae 368 112 30 8 Rubiaceae 616 110 18 9 Lamiaceae 435 95 22 10 Euphorbiaceae 523 94 18 B. TOTAL NO. OF SPECIES/NAMES OF FLOWERING TREES OF SIKKIM: (678 Species) Sl.No. Family, Genus & Species Height Altitude (in m) (in m) DILLENIACEAE 1. Dillenia indica L. 10-13 150-600 2. Dillenia pentagyna Roxb. 13-18 150-1500 MAGNOLIACEAE 3. Magnolia campbelli Hook. f. & Thomson 15-25 1000-2700 4. Magnolia globosa Hook. f. & Thomson 06-12 2300-3500 5. Magnolia hodgsonii (Hook. f. & Thomson) H. Keng 08-12 1000 6. Magnolia insignis Wall. 25-30 400-1500 7. Magnolia pterocarpa Roxb. 18-30 500 8. Michelia cathcartii Hook. f. & Thomson 15-25 1500-2000 9. Michelia champaca L. 03-05 10. Michelia doltsopa Buch. - Ham. ex DC. 16-25 1000-2500 11. Michelia glabra P.Pann. 10-15 900 12. Michelia kisopa Buch.-Ham. ex DC. 10-20 1400-1800 13. Michelia punduana Hook. f. & Thomson 12-03 1000-1600 14. Michelia velutina DC. -

Flavonoids: Classification, Biosynthesis and Chemical Ecology
Chapter 1 Flavonoids: Classification, Biosynthesis and Chemical Ecology Erica L. Santos, Beatriz Helena L.N. Sales Maia, Aurea P. Ferriani and Sirlei Dias Teixeira Additional information is available at the end of the chapter http://dx.doi.org/10.5772/67861 Abstract Flavonoids are natural products widely distributed in the plant kingdom and form one of the main classes of secondary metabolites. They display a large range of structures and ecological significance (e.g., such as the colored pigments in many flower petals), serve as chemotaxonomic marker compounds and have a variety of biological activi- ties. Therefore, they have been extensively investigated but the interest in them is still increasing. The topics that will be discussed in this chapter describe the regulation of- fla vonoid biosynthesis, the roles of flavonoids in flowers, fruits and roots and mechanisms involved in pollination and their specific functions in the plant. Keywords: flavonoids, biosynthesis, pollination, allelochemicals, chemical ecology 1. Introduction Flavonoids represent a highly diverse class of polyphenolic secondary metabolites, which are abundant in spermatophytes (seed-bearing vascular land plants: gymnosperms (cycades, conifers, ginkos and gnetophytes) and angiosperms) [1–3] but have also been reported from primitive taxa, such as bryophytes (nonvascular land plants, including liverworts, hornworts and mosses) [4, 5], pteridophytes (seedless vascular land plants, i.e., lycophytes, horsetails and all ferns) [6, 7] and algae [8, 9]. Overall, about 10,000 flavonoids have been recorded which represent the third largest group of natural products following the alkaloids (12,000) and terpenoids (30,000) [1, 10]. Flavonoids are essential constituents of the cells of all higher plants 11[ ]. -

Table of Contents
Forestry Department Food and Agriculture Organization of the United Nations FRA 2000 FOREST RESOURCES OF NEPAL COUNTRY REPORT Rome, 1999 Forest Resources Assessment Programme Working Paper 16 Rome 1999 The Forest Resources Assessment Programme Forests are crucial for the well-being of humanity. They provide foundations for life on earth through ecological functions, by regulating the climate and water resources, and by serving as habitats for plants and animals. Forests also furnish a wide range of essential goods such as wood, food, fodder and medicines, in addition to opportunities for recreation, spiritual renewal and other services. Today, forests are under pressure from expanding human populations, which frequently leads to the conversion or degradation of forests into unsustainable forms of land use. When forests are lost or severely degraded, their capacity to function as regulators of the environment is also lost, increasing flood and erosion hazards, reducing soil fertility, and contributing to the loss of plant and animal life. As a result, the sustainable provision of goods and services from forests is jeopardized. FAO, at the request of the member nations and the world community, regularly monitors the world’s forests through the Forest Resources Assessment Programme. The next report, the Global Forest Resources Assessment 2000 (FRA 2000), will review the forest situation by the end of the millennium. FRA 2000 will include country-level information based on existing forest inventory data, regional investigations of land- cover change processes, and a number of global studies focusing on the interaction between people and forests. The FRA 2000 report will be made public and distributed on the world wide web in the year 2000.