Genus-Specific Commensalism of the Galeommatoid Bivalve Koreamya Arcuata (A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chec List Bivalves of the São Sebastião Channel, North Coast Of
Check List 10(1): 97–105, 2014 © 2014 Check List and Authors Chec List ISSN 1809-127X (available at www.checklist.org.br) Journal of species lists and distribution Bivalves of the São Sebastião Channel, north coast of the PECIES S São Paulo State, Brazil OF Lenita de Freitas Tallarico 1*, Flávio Dias Passos 2, Fabrizio Marcondes Machado 3, Ariane Campos 1, ISTS 1 1,4 L Shirlei Maria Recco-Pimentel and Gisele Orlandi Introíni 1 Universidade Estadual de Campinas, Instituto de Biologia, Departamento de Biologia Estrutural e Funcional. R. Charles Darwin, s/n - Bloco N, Caixa Postal 6109. CEP 13083-863. Campinas, SP, Brazil. 2 Universidade Estadual de Campinas, Instituto de Biologia, Departamento de Biologia Animal. Rua Monteiro Lobato, 255, Caixa Postal 6109. CEP 13083-970. Campinas, SP, Brazil. 3 Programas de Pós-Graduação em Ecologia e Biologia Animal, Instituto de Biologia, Universidade Estadual de Campinas. R. Bertrand Russell, s/n, Caixa Postal 6109, CEP 13083-970. Campinas, SP, Brazil. 4 Universidade Federal de Ciências da Saúde de Porto Alegre, Departamento de Ciências Básicas da Saúde. R. Sarmento Leite, 245. CEP 90050-170. Porto Alegre, RS, Brazil. * Corresponding author. E-mail: [email protected] Abstract: The north coast of the São Paulo State, Brazil, presents great bivalve diversity, but knowledge about these organisms, especially species living subtidally, remains scarce. Based on collections made between 2010 and 2012, the present work provides a species list of bivalves inhabiting the intertidal and subtidal zones of the São Sebastião Channel. Altogether, 388 living specimens were collected, belonging to 52 species of 34 genera, grouped in 18 families. -

Biodiversità Ed Evoluzione
Allma Mater Studiiorum – Uniiversiità dii Bollogna DOTTORATO DI RICERCA IN BIODIVERSITÀ ED EVOLUZIONE Ciclo XXIII Settore/i scientifico-disciplinare/i di afferenza: BIO - 05 A MOLECULAR PHYLOGENY OF BIVALVE MOLLUSKS: ANCIENT RADIATIONS AND DIVERGENCES AS REVEALED BY MITOCHONDRIAL GENES Presentata da: Dr Federico Plazzi Coordinatore Dottorato Relatore Prof. Barbara Mantovani Dr Marco Passamonti Esame finale anno 2011 of all marine animals, the bivalve molluscs are the most perfectly adapted for life within soft substrata of sand and mud. Sir Charles Maurice Yonge INDEX p. 1..... FOREWORD p. 2..... Plan of the Thesis p. 3..... CHAPTER 1 – INTRODUCTION p. 3..... 1.1. BIVALVE MOLLUSKS: ZOOLOGY, PHYLOGENY, AND BEYOND p. 3..... The phylum Mollusca p. 4..... A survey of class Bivalvia p. 7..... The Opponobranchia: true ctenidia for a truly vexed issue p. 9..... The Autobranchia: between tenets and question marks p. 13..... Doubly Uniparental Inheritance p. 13..... The choice of the “right” molecular marker in bivalve phylogenetics p. 17..... 1.2. MOLECULAR EVOLUTION MODELS, MULTIGENE BAYESIAN ANALYSIS, AND PARTITION CHOICE p. 23..... CHAPTER 2 – TOWARDS A MOLECULAR PHYLOGENY OF MOLLUSKS: BIVALVES’ EARLY EVOLUTION AS REVEALED BY MITOCHONDRIAL GENES. p. 23..... 2.1. INTRODUCTION p. 28..... 2.2. MATERIALS AND METHODS p. 28..... Specimens’ collection and DNA extraction p. 30..... PCR amplification, cloning, and sequencing p. 30..... Sequence alignment p. 32..... Phylogenetic analyses p. 37..... Taxon sampling p. 39..... Dating p. 43..... 2.3. RESULTS p. 43..... Obtained sequences i p. 44..... Sequence analyses p. 45..... Taxon sampling p. 45..... Maximum Likelihood p. 47..... Bayesian Analyses p. 50..... Dating the tree p. -

NEAT Mollusca
NEAT (North East Atlantic Taxa): Scandinavian marine Mollusca Check-List compiled at TMBL (Tjärnö Marine Biological Laboratory) by: Hans G. Hansson 1994-02-02 / small revisions until February 1997, when it was published on Internet as a pdf file and then republished August 1998.. Citation suggested: Hansson, H.G. (Comp.), NEAT (North East Atlantic Taxa): Scandinavian marine Mollusca Check-List. Internet Ed., Aug. 1998. [http://www.tmbl.gu.se]. Denotations: (™) = Genotype @ = Associated to * = General note PHYLUM, CLASSIS, SUBCLASSIS, SUPERORDO, ORDO, SUBORDO, INFRAORDO, Superfamilia, Familia, Subfamilia, Genus & species N.B.: This is one of several preliminary check-lists, covering S Scandinavian marine animal (and partly marine protoctistan) taxa. Some financial support from (or via) NKMB (Nordiskt Kollegium för Marin Biologi), during the last years of the existence of this organization (until 1993), is thankfully acknowledged. The primary purpose of these checklists is to faciliate for everyone, trying to identify organisms from the area, to know which species that earlier have been encountered there, or in neighbouring areas. A secondary purpose is to faciliate for non-experts to put as correct names as possible on organisms, including names of authors and years of description. So far these checklists are very preliminary. Due to restricted access to literature there are (some known, and probably many unknown) omissions in the lists. Certainly also several errors may be found, especially regarding taxa like Plathelminthes and Nematoda, where the experience of the compiler is very rudimentary, or. e.g. Porifera, where, at least in certain families, taxonomic confusion seems to prevail. This is very much a small modernization of T. -
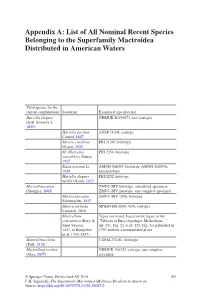
List of All Nominal Recent Species Belonging to the Superfamily Mactroidea Distributed in American Waters
Appendix A: List of All Nominal Recent Species Belonging to the Superfamily Mactroidea Distributed in American Waters Valid species (in the current combination) Synonym Examined type material Harvella elegans NHMUK 20190673, two syntypes (G.B. Sowerby I, 1825) Harvella pacifica ANSP 51308, syntype Conrad, 1867 Mactra estrellana PRI 21265, holotype Olsson, 1922 M. (Harvella) PRI 2354, holotype sanctiblasii Maury, 1925 Raeta maxima Li, AMNH 268093, lectotype; AMNH 268093a, 1930 paralectotype Harvella elegans PRI 2252, holotype tucilla Olsson, 1932 Mactrellona alata ZMUC-BIV, holotype, articulated specimen; (Spengler, 1802) ZMUC-BIV, paratype, one complete specimen Mactra laevigata ZMUC-BIV 1036, holotype Schumacher, 1817 Mactra carinata MNHN-IM-2000-7038, syntypes Lamarck, 1818 Mactrellona Types not found, based on the figure of the concentrica (Bory de “Tableau of Encyclopedique Methodique…” Saint Vincent, (pl. 251, Fig. 2a, b, pl. 252, Fig. 2c) published in 1827, in Bruguière 1797 without a nomenclatorial act et al. 1791–1827) Mactrellona clisia USNM 271481, holotype (Dall, 1915) Mactrellona exoleta NHMUK 196327, syntype, one complete (Gray, 1837) specimen © Springer Nature Switzerland AG 2019 103 J. H. Signorelli, The Superfamily Mactroidea (Mollusca:Bivalvia) in American Waters, https://doi.org/10.1007/978-3-030-29097-9 104 Appendix A: List of All Nominal Recent Species Belonging to the Superfamily… Valid species (in the current combination) Synonym Examined type material Lutraria ventricosa MCZ 169451, holotype; MCZ 169452, paratype; -

Worm-Riding Clam: Description of Montacutona Sigalionidcola Sp. Nov. (Bivalvia: Heterodonta: Galeommatidae) from Japan and Its Phylogenetic Position
Zootaxa 4652 (3): 473–486 ISSN 1175-5326 (print edition) https://www.mapress.com/j/zt/ Article ZOOTAXA Copyright © 2019 Magnolia Press ISSN 1175-5334 (online edition) https://doi.org/10.11646/zootaxa.4652.3.4 http://zoobank.org/urn:lsid:zoobank.org:pub:D1823BBA-EDC4-4A1D-84C1-27BD4F002CE0 Worm-riding clam: description of Montacutona sigalionidcola sp. nov. (Bivalvia: Heterodonta: Galeommatidae) from Japan and its phylogenetic position RYUTARO GOTO1,3 & MAKOTO TANAKA2 1Seto Marine Biological Laboratory, Field Science Education and Research Center, Kyoto University, 459 Shirahama, Nishimuro, Wakayama 649-2211, Japan 21060–113 Sangodai, Kushimoto, Higashimuro, Wakayama 649-3510, Japan 3Corresponging author. E-mail: [email protected] Abstract A new galeommatid bivalve, Montacutona sigalionidcola sp. nov., is described from an intertidal flat in the southern end of the Kii Peninsula, Honshu Island, Japan. Unlike other members of the genus, this species is a commensal with the burrowing scale worm Pelogenia zeylanica (Willey) (Annelida: Sigalionidae) that lives in fine sand sediments. Specimens were always found attached to the dorsal surface of the anterior end of the host body. This species has a ligament lithodesma between diverging hinge teeth, which is characteristic of Montacutona Yamamoto & Habe. However, it is morphologically distinguished from the other members of this genus in having elongate-oval shells with small gape at the posteroventral margin and lacking an outer demibranch. Molecular phylogenetic analysis based on the four-gene combined dataset (18S + 28S + H3 + COI) indicated that this species is monophyletic with Montacutona, Nipponomontacuta Yamamoto & Habe and Koreamya Lützen, Hong & Yamashita, which are commensals with sea anemones or Lingula brachiopods. -

Mollusca: Bivalvia
AR-1580 11 MOLLUSCA: BIVALVIA Robert F. McMahon Arthur E. Bogan Department of Biology North Carolina State Museum Box 19498 of Natural Sciences The University of Texas at Arlington Research Laboratory Arlington, TX 76019 4301 Ready Creek Road Raleigh, NC 27607 I. Introduction A. Collecting II. Anatomy and Physiology B. Preparation for Identification A. External Morphology C. Rearing Freshwater Bivalves B. Organ-System Function V. Identification of the Freshwater Bivalves C. Environmental and Comparative of North America Physiology A. Taxonomic Key to the Superfamilies of III. Ecology and Evolution Freshwater Bivalvia A. Diversity and Distribution B. Taxonomic Key to Genera of B. Reproduction and Life History Freshwater Corbiculacea C. Ecological Interactions C. Taxonomic Key to the Genera of D. Evolutionary Relationships Freshwater Unionoidea IV. Collecting, Preparation for Identification, Literature Cited and Rearing I. INTRODUCTION ament uniting the calcareous valves (Fig. 1). The hinge lig- ament is external in all freshwater bivalves. Its elasticity North American (NA) freshwater bivalve molluscs opens the valves while the anterior and posterior shell ad- (class Bivalvia) fall in the subclasses Paleoheterodonta (Su- ductor muscles (Fig. 2) run between the valves and close perfamily Unionoidea) and Heterodonta (Superfamilies them in opposition to the hinge ligament which opens Corbiculoidea and Dreissenoidea). They have enlarged them on adductor muscle relaxation. gills with elongated, ciliated filaments for suspension feed- The mantle lobes and shell completely enclose the ing on plankton, algae, bacteria, and microdetritus. The bivalve body, resulting in cephalic sensory structures be- mantle tissue underlying and secreting the shell forms a coming vestigial or lost. Instead, external sensory struc- pair of lateral, dorsally connected lobes. -

Comparative Shell Microstructure of Two Species of Tropical Laternulid
Comparative Shell Microstructure of Two Species of Tropical Laternulid Bivalves from Kungkrabaen Bay, Thailand with After-Thoughts on Laternulid Taxonomy Author(s): Robert S. Prezant, Rebecca M. Shell, and Laying Wu Source: American Malacological Bulletin, 33(1):22-33. Published By: American Malacological Society DOI: http://dx.doi.org/10.4003/006.033.0112 URL: http://www.bioone.org/doi/full/10.4003/006.033.0112 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. AMERICAN MALACOLOGICAL BULLETIN BULLETIN AMERICAN MALACOLOGICAL OLUME UMBER American Malacological Bulletin 33(1), 13 March 2015 V 33, N 1 13 March 2015 Independent Papers Functional morphology of Neolepton. Brian Morton . .1 Comparative shell microstructure of two species of tropical laternulid bivalves from Kungkrabaen Bay, Thailand with after-thoughts on laternulid taxonomy. Robert S. Prezant, Rebecca M. Shell, and Laying Wu . -

Unorthodox Features in Two Venerid Bivalves with Doubly Uniparental
Unorthodox features in two venerid bivalves with doubly uniparental inheritance of mitochondria Charlotte Capt, Karim Bouvet, Davide Guerra, Brent Robicheau, Donald Stewart, Eric Pante, Sophie Breton To cite this version: Charlotte Capt, Karim Bouvet, Davide Guerra, Brent Robicheau, Donald Stewart, et al.. Unortho- dox features in two venerid bivalves with doubly uniparental inheritance of mitochondria. Scientific Reports, Nature Publishing Group, 2020, 10 (1), 10.1038/s41598-020-57975-y. hal-02499562 HAL Id: hal-02499562 https://hal.archives-ouvertes.fr/hal-02499562 Submitted on 8 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License www.nature.com/scientificreports OPEN Unorthodox features in two venerid bivalves with doubly uniparental inheritance of mitochondria Charlotte Capt1*, Karim Bouvet1, Davide Guerra1, Brent M. Robicheau2, Donald T. Stewart3, Eric Pante4,5 & Sophie Breton1,5* In animals, strictly maternal inheritance (SMI) of mitochondria is the rule, but one exception (doubly uniparental inheritance or DUI), marked by the transmission of sex-specifc mitogenomes, has been reported in bivalves. Associated with DUI is a frequent modifcation of the mitochondrial cox2 gene, as well as additional sex-specifc mitochondrial genes not involved in oxidative phosphorylation.