Cloning, Characterization and Expression of CDK5RAP1 V3 and CDK5RAP1 V4, Two Novel Splice Variants of Human CDK5RAP1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cdk5rap1 (NM 025876) Mouse Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for MR209175 Cdk5rap1 (NM_025876) Mouse Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: Cdk5rap1 (NM_025876) Mouse Tagged ORF Clone Tag: Myc-DDK Symbol: Cdk5rap1 Synonyms: 2310066P17Rik Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 4 Cdk5rap1 (NM_025876) Mouse Tagged ORF Clone – MR209175 ORF Nucleotide >MR209175 ORF sequence Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGCATCCTTTACGGTGTGTCCTCCAAGTACAGAGGTTGTCAGCACCATTCACCTCCATGTGCTGGGTGT TGCTTAGGACCTGCAGAGCACAAAGCAGTGTGTCCAGCACTCCTTGTCCCAGTCCAGAGGCGAAGAGCTC AGAAGCTCAGAAGGACTTCAGCTCCAGGCTAGCCACTGGACCGACTTTTCAGCATTTTTTAAGAAGTGCT TCAGTTCCTCAAGAGAAACCATCTTCTCCAGAGGTGGAGGACCCACCTCCCTATCTCTCGGGGGATGAAC TTCTAGGAAGGCAGAGAAAAGTCTACCTCGAGACCTATGGCTGTCAGATGAACGTGAACGACACAGAGAT AGCCTGGTCCATCTTACAGAAGAGTGGCTACCTTCGGACCAGCAACCTCCAAGAGGCTGATGTGATCCTT CTTGTCACGTGTTCTATCAGGGAGAAGGCCGAGCAGACCATCTGGAACCGCTTACATCAGCTCAAAGTCC TGAAGACAAAGCGGCCACGCTCTCGAGTACCTCTGAGGATTGGGATTCTAGGCTGCATGGCTGAGAGGCT GAAGGGAGAGATCCTCAACAGGGAGAAGATGGTAGATCTCTTGGCTGGTCCAGACGCCTACCGAGACCTT -
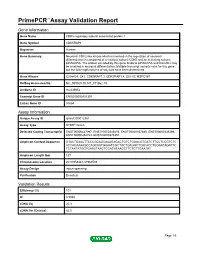
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name CDK5 regulatory subunit associated protein 1 Gene Symbol CDK5RAP1 Organism Human Gene Summary Neuronal CDC2-like kinase which is involved in the regulation of neuronal differentiation is composed of a catalytic subunit CDK5 and an activating subunit p25NCK5A. The protein encoded by this gene binds to p25NCK5A and therefore may be involved in neuronal differentiation. Multiple transcript variants exist for this gene but the full-length natures of only two have been determined. Gene Aliases C20orf34, C42, CDK5RAP1.3, CDK5RAP1.4, CGI-05, HSPC167 RefSeq Accession No. NC_000020.10, NT_011362.10 UniGene ID Hs.435952 Ensembl Gene ID ENSG00000101391 Entrez Gene ID 51654 Assay Information Unique Assay ID qHsaCID0012381 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000427097, ENST00000346416, ENST00000357886, ENST00000339269, ENST00000452723, ENST00000375351 Amplicon Context Sequence GTACTGAACTTCCCGGAGCAAAGAGACTGTCTGGACGTGATCTTCCTCCGTCTC ACCACAAAAGCCAGCAATGAAATCGCTGCTGAGGCTCACACCTGGAATAGATTC TCTAATATGGTGAACTAACTCCACATAAGCTTCTCTTGAATAT Amplicon Length (bp) 121 Chromosome Location 20:31958341-31960503 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 101 R2 0.9988 cDNA Cq 20.3 cDNA Tm (Celsius) 82.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report CDK5RAP1, Human Amplification Plot Amplification -

Chromosomal Microarray Analysis in Turkish Patients with Unexplained Developmental Delay and Intellectual Developmental Disorders
177 Arch Neuropsychitry 2020;57:177−191 RESEARCH ARTICLE https://doi.org/10.29399/npa.24890 Chromosomal Microarray Analysis in Turkish Patients with Unexplained Developmental Delay and Intellectual Developmental Disorders Hakan GÜRKAN1 , Emine İkbal ATLI1 , Engin ATLI1 , Leyla BOZATLI2 , Mengühan ARAZ ALTAY2 , Sinem YALÇINTEPE1 , Yasemin ÖZEN1 , Damla EKER1 , Çisem AKURUT1 , Selma DEMİR1 , Işık GÖRKER2 1Faculty of Medicine, Department of Medical Genetics, Edirne, Trakya University, Edirne, Turkey 2Faculty of Medicine, Department of Child and Adolescent Psychiatry, Trakya University, Edirne, Turkey ABSTRACT Introduction: Aneuploids, copy number variations (CNVs), and single in 39 (39/123=31.7%) patients. Twelve CNV variant of unknown nucleotide variants in specific genes are the main genetic causes of significance (VUS) (9.75%) patients and 7 CNV benign (5.69%) patients developmental delay (DD) and intellectual disability disorder (IDD). were reported. In 6 patients, one or more pathogenic CNVs were These genetic changes can be detected using chromosome analysis, determined. Therefore, the diagnostic efficiency of CMA was found to chromosomal microarray (CMA), and next-generation DNA sequencing be 31.7% (39/123). techniques. Therefore; In this study, we aimed to investigate the Conclusion: Today, genetic analysis is still not part of the routine in the importance of CMA in determining the genomic etiology of unexplained evaluation of IDD patients who present to psychiatry clinics. A genetic DD and IDD in 123 patients. diagnosis from CMA can eliminate genetic question marks and thus Method: For 123 patients, chromosome analysis, DNA fragment analysis alter the clinical management of patients. Approximately one-third and microarray were performed. Conventional G-band karyotype of the positive CMA findings are clinically intervenable. -

CDK5RAP1 Lentiviral Activation Particles (M): Sc-426299-LAC
SANTA CRUZ BIOTECHNOLOGY, INC. CDK5RAP1 Lentiviral Activation Particles (m): sc-426299-LAC BACKGROUND APPLICATIONS The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and Either CDK5RAP1 Lentiviral Activation Particles (m) or CDK5RAP1 Lentiviral CRISPR-associated protein (Cas9) system is an adaptive immune response Activation Particles (m2) is recommended for gene activation in mouse cells. defense mechanism used by archea and bacteria for the degradation of for- eign genetic material. This mechanism can be repurposed for other functions, SUPPORT REAGENTS including genomic engineering for mammalian systems, such as gene knock- For optimal reaction efficiency with Lentiviral Activation Particle products, out (KO) (1,2) and gene activation (3-5). CRISPR Activation Plasmid products Polybrene®: sc-134220 (10 mg/ml) is recommended. Hygromycin B solution: enable the identification and upregulation of specific genes by utilizing a sc-29067 (1 g), Blasticidin S HCl solution: sc-495389 (1 ml) and Puromycin D10A and N863A deactivated Cas9 (dCas9) nuclease fused to a VP64 acti- dihydrochloride: sc-108071 (25 mg) are recommended for selection. Control vation domain, in conjunction with sgRNA (MS2), a target-specific sgRNA Lentiviral Activation Particles: sc-437282 (200 µl) negative control is also engineered to bind the MS2-P65-HSF1 fusion protein (5). This synergistic available. activation mediator (SAM) transcription activation system* provides a robust system to maximize the activation of endogenous gene expression (5). GENE EXPRESSION MONITORING REFERENCES CDK5RAP1 (D-1): sc-398764 is recommended as a control antibody for moni- toring of Cdk5rap1 (mouse) gene expression prior to and after activation by 1. Cong, L., et al. 2013. Multiplex genome engineering using CRISPR/Cas Western blotting (starting dilution 1:200, dilution range 1:100-1:1000) or systems. -

Genomic Landscape of Paediatric Adrenocortical Tumours
ARTICLE Received 5 Aug 2014 | Accepted 16 Jan 2015 | Published 6 Mar 2015 DOI: 10.1038/ncomms7302 Genomic landscape of paediatric adrenocortical tumours Emilia M. Pinto1,*, Xiang Chen2,*, John Easton2, David Finkelstein2, Zhifa Liu3, Stanley Pounds3, Carlos Rodriguez-Galindo4, Troy C. Lund5, Elaine R. Mardis6,7,8, Richard K. Wilson6,7,9, Kristy Boggs2, Donald Yergeau2, Jinjun Cheng2, Heather L. Mulder2, Jayanthi Manne2, Jesse Jenkins10, Maria J. Mastellaro11, Bonald C. Figueiredo12, Michael A. Dyer13, Alberto Pappo14, Jinghui Zhang2, James R. Downing10, Raul C. Ribeiro14,* & Gerard P. Zambetti1,* Paediatric adrenocortical carcinoma is a rare malignancy with poor prognosis. Here we analyse 37 adrenocortical tumours (ACTs) by whole-genome, whole-exome and/or transcriptome sequencing. Most cases (91%) show loss of heterozygosity (LOH) of chromosome 11p, with uniform selection against the maternal chromosome. IGF2 on chromosome 11p is overexpressed in 100% of the tumours. TP53 mutations and chromosome 17 LOH with selection against wild-type TP53 are observed in 28 ACTs (76%). Chromosomes 11p and 17 undergo copy-neutral LOH early during tumorigenesis, suggesting tumour-driver events. Additional genetic alterations include recurrent somatic mutations in ATRX and CTNNB1 and integration of human herpesvirus-6 in chromosome 11p. A dismal outcome is predicted by concomitant TP53 and ATRX mutations and associated genomic abnormalities, including massive structural variations and frequent background mutations. Collectively, these findings demonstrate the nature, timing and potential prognostic significance of key genetic alterations in paediatric ACT and outline a hypothetical model of paediatric adrenocortical tumorigenesis. 1 Department of Biochemistry, St Jude Children’s Research Hospital, Memphis, Tennessee 38105, USA. 2 Department of Computational Biology and Bioinformatics, St Jude Children’s Research Hospital, Memphis, Tennessee 38105, USA. -

PDF Download
CDK5 Activator-binding C48 Polyclonal Antibody Catalog No : YT0837 Reactivity : Human,Rat,Mouse, Applications : IHC-p,IF(paraffin section),ELISA Gene Name : CDK5RAP2 Protein Name : CDK5 regulatory subunit-associated protein 2 Human Gene Id : 55755 Human Swiss Prot Q96SN8 No : Mouse Swiss Prot Q8K389 No : Immunogen : The antiserum was produced against synthesized peptide derived from human CDK5RAP2. AA range:251-300 Specificity : CDK5 Activator-binding C48 Polyclonal Antibody detects endogenous levels of CDK5 Activator-binding C48 protein. Formulation : Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide. Source : Rabbit Dilution : Immunohistochemistry: 1/100 - 1/300. ELISA: 1/20000. Not yet tested in other applications. Purification : The antibody was affinity-purified from rabbit antiserum by affinity- chromatography using epitope-specific immunogen. Concentration : 1 mg/ml Storage Stability : -20°C/1 year Molecularweight : 215024 1 / 2 Background : CDK5 regulatory subunit associated protein 2(CDK5RAP2) Homo sapiens This gene encodes a regulator of CDK5 (cyclin-dependent kinase 5) activity. The protein encoded by this gene is localized to the centrosome and Golgi complex, interacts with CDK5R1 and pericentrin (PCNT), plays a role in centriole engagement and microtubule nucleation, and has been linked to primary microcephaly and Alzheimer's disease. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2013], Function : disease:Defects in CDK5RAP2 are the cause of primary microcephaly autosomal recessive type 3 (MCPH3) [MIM:604804]. Microcephaly is defined as a head circumference more than 3 standard deviations below the age-related mean. Brain weight is markedly reduced and the cerebral cortex is disproportionately small. -

Identification of Novel Regulatory Genes in Acetaminophen
IDENTIFICATION OF NOVEL REGULATORY GENES IN ACETAMINOPHEN INDUCED HEPATOCYTE TOXICITY BY A GENOME-WIDE CRISPR/CAS9 SCREEN A THESIS IN Cell Biology and Biophysics and Bioinformatics Presented to the Faculty of the University of Missouri-Kansas City in partial fulfillment of the requirements for the degree DOCTOR OF PHILOSOPHY By KATHERINE ANNE SHORTT B.S, Indiana University, Bloomington, 2011 M.S, University of Missouri, Kansas City, 2014 Kansas City, Missouri 2018 © 2018 Katherine Shortt All Rights Reserved IDENTIFICATION OF NOVEL REGULATORY GENES IN ACETAMINOPHEN INDUCED HEPATOCYTE TOXICITY BY A GENOME-WIDE CRISPR/CAS9 SCREEN Katherine Anne Shortt, Candidate for the Doctor of Philosophy degree, University of Missouri-Kansas City, 2018 ABSTRACT Acetaminophen (APAP) is a commonly used analgesic responsible for over 56,000 overdose-related emergency room visits annually. A long asymptomatic period and limited treatment options result in a high rate of liver failure, generally resulting in either organ transplant or mortality. The underlying molecular mechanisms of injury are not well understood and effective therapy is limited. Identification of previously unknown genetic risk factors would provide new mechanistic insights and new therapeutic targets for APAP induced hepatocyte toxicity or liver injury. This study used a genome-wide CRISPR/Cas9 screen to evaluate genes that are protective against or cause susceptibility to APAP-induced liver injury. HuH7 human hepatocellular carcinoma cells containing CRISPR/Cas9 gene knockouts were treated with 15mM APAP for 30 minutes to 4 days. A gene expression profile was developed based on the 1) top screening hits, 2) overlap with gene expression data of APAP overdosed human patients, and 3) biological interpretation including assessment of known and suspected iii APAP-associated genes and their therapeutic potential, predicted affected biological pathways, and functionally validated candidate genes. -

Kinome Expression Profiling to Target New Therapeutic Avenues in Multiple Myeloma
Plasma Cell DIsorders SUPPLEMENTARY APPENDIX Kinome expression profiling to target new therapeutic avenues in multiple myeloma Hugues de Boussac, 1 Angélique Bruyer, 1 Michel Jourdan, 1 Anke Maes, 2 Nicolas Robert, 3 Claire Gourzones, 1 Laure Vincent, 4 Anja Seckinger, 5,6 Guillaume Cartron, 4,7,8 Dirk Hose, 5,6 Elke De Bruyne, 2 Alboukadel Kassambara, 1 Philippe Pasero 1 and Jérôme Moreaux 1,3,8 1IGH, CNRS, Université de Montpellier, Montpellier, France; 2Department of Hematology and Immunology, Myeloma Center Brussels, Vrije Universiteit Brussel, Brussels, Belgium; 3CHU Montpellier, Laboratory for Monitoring Innovative Therapies, Department of Biologi - cal Hematology, Montpellier, France; 4CHU Montpellier, Department of Clinical Hematology, Montpellier, France; 5Medizinische Klinik und Poliklinik V, Universitätsklinikum Heidelberg, Heidelberg, Germany; 6Nationales Centrum für Tumorerkrankungen, Heidelberg , Ger - many; 7Université de Montpellier, UMR CNRS 5235, Montpellier, France and 8 Université de Montpellier, UFR de Médecine, Montpel - lier, France ©2020 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2018.208306 Received: October 5, 2018. Accepted: July 5, 2019. Pre-published: July 9, 2019. Correspondence: JEROME MOREAUX - [email protected] Supplementary experiment procedures Kinome Index A list of 661 genes of kinases or kinases related have been extracted from literature9, and challenged in the HM cohort for OS prognostic values The prognostic value of each of the genes was computed using maximally selected rank test from R package MaxStat. After Benjamini Hochberg multiple testing correction a list of 104 significant prognostic genes has been extracted. This second list has then been challenged for similar prognosis value in the UAMS-TT2 validation cohort. -

A High-Density Human Mitochondrial Proximity Interaction Network
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.01.020479; this version posted April 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. A high-density human mitochondrial proximity interaction network Hana Antonicka1,2, Zhen-Yuan Lin3, Alexandre Janer1,2, Woranontee Weraarpachai1,5, Anne- Claude Gingras3,4,*, Eric A. Shoubridge1,2* 1 Montreal Neurological Institute, McGill University, Montreal, QC, Canada 2 Department of Human Genetics, McGill University, Montreal, QC, Canada 3 Lunenfeld-Tanenbaum Research Institute, Toronto, ON, Canada 4 Department of Molecular Genetics, University of Toronto, Toronto, ON, Canada 5 Department of Biochemistry, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand Data deposition: Mass spectrometry data have been deposited in the Mass spectrometry Interactive Virtual Environment (MassIVE, http://massive.ucsd.edu). * corresponding authors Correspondence: e-mail: [email protected] Phone: (514) 398-1997 Fax: (514) 398-1509 e-mail: [email protected] Phone: (416) 586-5027 Fax: (416) 586-8869 Summary We used BioID, a proximity-dependent biotinylation assay, to interrogate 100 mitochondrial baits from all mitochondrial sub-compartments to create a high resolution human mitochondrial proximity interaction network. We identified 1465 proteins, producing 15626 unique high confidence proximity interactions. Of these, 528 proteins were previously annotated as mitochondrial, nearly half of the mitochondrial proteome defined by Mitocarta 2.0. Bait-bait analysis showed a clear separation of mitochondrial compartments, and correlation analysis among preys across all baits allowed us to identify functional clusters involved in diverse mitochondrial functions, and to assign uncharacterized proteins to specific modules. -
Inhibition of Cdk5 Promotes Β-Cell Differentiation from Ductal
58 Diabetes Volume 67, January 2018 Inhibition of Cdk5 Promotes b-Cell Differentiation From Ductal Progenitors Ka-Cheuk Liu,1 Gunter Leuckx,2 Daisuke Sakano,3 Philip A. Seymour,4 Charlotte L. Mattsson,1 Linn Rautio,1 Willem Staels,2 Yannick Verdonck,2 Palle Serup,4 Shoen Kume,3 Harry Heimberg,2 and Olov Andersson1 Diabetes 2018;67:58–70 | https://doi.org/10.2337/db16-1587 Inhibition of notch signaling is known to induce differentiation have shown that pancreatic ductal ligation (PDL) or over- of endocrine cells in zebrafish and mouse. After performing expression of the transcription factor Pax4 in a-cells in- an unbiased in vivo screen of ∼2,200 small molecules in duces neogenesis of endocrine cells originating from the zebrafish, we identified an inhibitor of Cdk5 (roscovitine), pancreatic duct (8–10). In humans, acinar-associated neo- which potentiated the formation of b-cells along the intra- genesis was promoted in obese donors without diabetes pancreatic duct during concurrent inhibition of notch sig- whereas duct-associated neogenesis was increased in both fi naling. We con rmed and characterized the effect with a lean and obese donors with type 2 diabetes (6). Despite more selective Cdk5 inhibitor, (R)-DRF053, which spe- being widely reported, some studies show that neogenesis fi b ci cally increased the number of duct-derived -cells of b-cells rarely occurs or even does not happen in certain without affecting their proliferation. By duct-specificover- experimental conditions (11,12). This discrepancy suggests expression of the endogenous Cdk5 inhibitors Cdk5rap1 that b-cell neogenesis is a precisely controlled event and it or Cdkal1 (which previously have been linked to diabetes in is likely limited endogenously. -

Mouse Endogenous Retroviruses Can Trigger Premature Transcriptional Termination at a Distance
Downloaded from genome.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Research Mouse endogenous retroviruses can trigger premature transcriptional termination at a distance Jingfeng Li,1 Keiko Akagi,1 Yongjun Hu,2 Anna L. Trivett,3 Christopher J.W. Hlynialuk,1 Deborah A. Swing,4 Natalia Volfovsky,5 Tamara C. Morgan,6 Yelena Golubeva,6 Robert M. Stephens,5 David E. Smith,2 and David E. Symer1,7,8 1Human Cancer Genetics Program and Department of Molecular Virology, Immunology and Medical Genetics, The Ohio State University Comprehensive Cancer Center, Columbus, Ohio 43210, USA; 2Department of Pharmaceutical Sciences, University of Michigan, Ann Arbor, Michigan 48109, USA; 3Laboratory of Molecular Immunoregulation and 4Mouse Cancer Genetics Program, National Cancer Institute, Frederick, Maryland 21702, USA; 5Advanced Biomedical Computing Center, Information Systems Program and 6Histotechnology Laboratory, SAIC-Frederick, Inc., National Cancer Institute, Frederick, Maryland 21702, USA; 7Department of Internal Medicine and Department of Biomedical Informatics, The Ohio State University Comprehensive Cancer Center, Columbus, Ohio 43210, USA Endogenous retrotransposons have caused extensive genomic variation within mammalian species, but the functional implications of such mobilization are mostly unknown. We mapped thousands of endogenous retrovirus (ERV) germline integrants in highly divergent, previously unsequenced mouse lineages, facilitating a comparison of gene expression in the presence or absence of -

A Point Mutation in the Ncr1 S
A point mutation in the Ncr1 signal peptide impairs the development of innate lymphoid cell subsets. Francisca F. Almeida, Sara Tognarelli, Antoine Marcais, Andrew J. Kueh, Miriam E. Friede, Yang Liao, Simon N. Willis, Kylie Luong, Fabrice Faure, Francois E. Mercier, et al. To cite this version: Francisca F. Almeida, Sara Tognarelli, Antoine Marcais, Andrew J. Kueh, Miriam E. Friede, et al.. A point mutation in the Ncr1 signal peptide impairs the development of innate lymphoid cell subsets.. OncoImmunology, Taylor & Francis, 2018, 7 (10), pp.e1475875. 10.1080/2162402X.2018.1475875. hal-02086847 HAL Id: hal-02086847 https://hal-amu.archives-ouvertes.fr/hal-02086847 Submitted on 1 Apr 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - NoDerivatives| 4.0 International License OncoImmunology ISSN: (Print) 2162-402X (Online) Journal homepage: https://www.tandfonline.com/loi/koni20 A point mutation in the Ncr1 signal peptide impairs the development of innate lymphoid cell subsets Francisca F. Almeida, Sara Tognarelli, Antoine Marçais, Andrew J. Kueh, Miriam E. Friede, Yang Liao, Simon N. Willis, Kylie Luong, Fabrice Faure, Francois E. Mercier, Justine Galluso, Matthew Firth, Emilie Narni-Mancinelli, Bushra Rais, David T.