Human Mitochondrial RNA Processing and Modifications
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Establishing the Pathogenicity of Novel Mitochondrial DNA Sequence Variations: a Cell and Molecular Biology Approach
Mafalda Rita Avó Bacalhau Establishing the Pathogenicity of Novel Mitochondrial DNA Sequence Variations: a Cell and Molecular Biology Approach Tese de doutoramento do Programa de Doutoramento em Ciências da Saúde, ramo de Ciências Biomédicas, orientada pela Professora Doutora Maria Manuela Monteiro Grazina e co-orientada pelo Professor Doutor Henrique Manuel Paixão dos Santos Girão e pela Professora Doutora Lee-Jun C. Wong e apresentada à Faculdade de Medicina da Universidade de Coimbra Julho 2017 Faculty of Medicine Establishing the pathogenicity of novel mitochondrial DNA sequence variations: a cell and molecular biology approach Mafalda Rita Avó Bacalhau Tese de doutoramento do programa em Ciências da Saúde, ramo de Ciências Biomédicas, realizada sob a orientação científica da Professora Doutora Maria Manuela Monteiro Grazina; e co-orientação do Professor Doutor Henrique Manuel Paixão dos Santos Girão e da Professora Doutora Lee-Jun C. Wong, apresentada à Faculdade de Medicina da Universidade de Coimbra. Julho, 2017 Copyright© Mafalda Bacalhau e Manuela Grazina, 2017 Esta cópia da tese é fornecida na condição de que quem a consulta reconhece que os direitos de autor são pertença do autor da tese e do orientador científico e que nenhuma citação ou informação obtida a partir dela pode ser publicada sem a referência apropriada e autorização. This copy of the thesis has been supplied on the condition that anyone who consults it recognizes that its copyright belongs to its author and scientific supervisor and that no quotation from the -

Mt-Atp8 Gene in the Conplastic Mouse Strain C57BL/6J-Mtfvb/NJ on the Mitochondrial Function and Consequent Alterations to Metabolic and Immunological Phenotypes
From the Lübeck Institute of Experimental Dermatology of the University of Lübeck Director: Prof. Dr. Saleh M. Ibrahim Interplay of mtDNA, metabolism and microbiota in the pathogenesis of AIBD Dissertation for Fulfillment of Requirements for the Doctoral Degree of the University of Lübeck from the Department of Natural Sciences Submitted by Paul Schilf from Rostock Lübeck, 2016 First referee: Prof. Dr. Saleh M. Ibrahim Second referee: Prof. Dr. Stephan Anemüller Chairman: Prof. Dr. Rainer Duden Date of oral examination: 30.03.2017 Approved for printing: Lübeck, 06.04.2017 Ich versichere, dass ich die Dissertation ohne fremde Hilfe angefertigt und keine anderen als die angegebenen Hilfsmittel verwendet habe. Weder vorher noch gleichzeitig habe ich andernorts einen Zulassungsantrag gestellt oder diese Dissertation vorgelegt. ABSTRACT Mitochondria are critical in the regulation of cellular metabolism and influence signaling processes and inflammatory responses. Mitochondrial DNA mutations and mitochondrial dysfunction are known to cause a wide range of pathological conditions and are associated with various immune diseases. The findings in this work describe the effect of a mutation in the mitochondrially encoded mt-Atp8 gene in the conplastic mouse strain C57BL/6J-mtFVB/NJ on the mitochondrial function and consequent alterations to metabolic and immunological phenotypes. This work provides insights into the mutation-induced cellular adaptations that influence the inflammatory milieu and shape pathological processes, in particular focusing on autoimmune bullous diseases, which have recently been reported to be associated with mtDNA polymorphisms in the human MT-ATP8 gene. The mt-Atp8 mutation diminishes the assembly of the ATP synthase complex into multimers and decreases mitochondrial respiration, affects generation of reactive oxygen species thus leading to a shift in the metabolic balance and reduction in the energy state of the cell as indicated by the ratio ATP to ADP. -

Mutation of the Fumarase Gene in Two Siblings with Progressive Encephalopathy and Fumarase Deficiency T
Mutation of the Fumarase Gene in Two Siblings with Progressive Encephalopathy and Fumarase Deficiency T. Bourgeron,* D. Chretien,* J. Poggi-Bach, S. Doonan,' D. Rabier,* P. Letouze,I A. Munnich,* A. R6tig,* P. Landneu,* and P. Rustin* *Unite de Recherches sur les Handicaps Genetiques de l'Enfant, INSERM U393, Departement de Pediatrie et Departement de Biochimie, H6pital des Enfants-Malades, 149, rue de Sevres, 75743 Paris Cedex 15, France; tDepartement de Pediatrie, Service de Neurologie et Laboratoire de Biochimie, Hopital du Kremlin-Bicetre, France; IFaculty ofScience, University ofEast-London, UK; and IService de Pediatrie, Hopital de Dreux, France Abstract chondrial enzyme (7). Human tissue fumarase is almost We report an inborn error of the tricarboxylic acid cycle, fu- equally distributed between the mitochondria, where the en- marase deficiency, in two siblings born to first cousin parents. zyme catalyzes the reversible hydration of fumarate to malate They presented with progressive encephalopathy, dystonia, as a part ofthe tricarboxylic acid cycle, and the cytosol, where it leucopenia, and neutropenia. Elevation oflactate in the cerebro- is involved in the metabolism of the fumarate released by the spinal fluid and high fumarate excretion in the urine led us to urea cycle. The two isoenzymes have quite homologous struc- investigate the activities of the respiratory chain and of the tures. In rat liver, they differ only by the acetylation of the Krebs cycle, and to finally identify fumarase deficiency in these NH2-terminal amino acid of the cytosolic form (8). In all spe- two children. The deficiency was profound and present in all cies investigated so far, the two isoenzymes have been found to tissues investigated, affecting the cytosolic and the mitochon- be encoded by a single gene (9,10). -

Unravelling the Cellular Origin and Clinical Prognostic Markers of Infant
Published Ahead of Print on January 24, 2019, as doi:10.3324/haematol.2018.206375. Copyright 2019 Ferrata Storti Foundation. Unravelling the cellular origin and clinical prognostic markers of infant B-cell acute lymphoblastic leukemia using genome-wide analysis by Antonio Agraz-Doblas, Clara Bueno, Rachael Bashford-Rogers, Anindita Roy, Pauline Schneider, Michela Bardini, Paola Ballerini, Gianni Cazzaniga, Thaidy Moreno, Carlos Revilla, Marta Gut, Maria G Valsecchi, Irene Roberts, Rob Pieters, Paola De Lorenzo, Ignacio Varela, Pablo Menendez, and Ronald W Stam Haematologica 2019 [Epub ahead of print] Citation: Antonio Agraz-Doblas, Clara Bueno, Rachael Bashford-Rogers, Anindita Roy, Pauline Schneider, Michela Bardini, Paola Ballerini, Gianni Cazzaniga, Thaidy Moreno, Carlos Revilla, Marta Gut, Maria G Valsecchi, Irene Roberts, Rob Pieters, Paola De Lorenzo, Ignacio Varela, Pablo Menendez, and Ronald W Stam. Unravelling the cellular origin and clinical prognostic markers of infant B-cell acute lymphoblastic leukemia using genome-wide analysis Haematologica. 2019; 104:xxx doi:10.3324/haematol.2018.206375 Publisher's Disclaimer. E-publishing ahead of print is increasingly important for the rapid dissemination of science. Haematologica is, therefore, E-publishing PDF files of an early version of manuscripts that have completed a regular peer review and have been accepted for publication. E-publishing of this PDF file has been approved by the authors. After having E-published Ahead of Print, manuscripts will then undergo technical and English editing, typesetting, proof correction and be presented for the authors' final approval; the final version of the manuscript will then appear in print on a regular issue of the journal. All legal disclaimers that apply to the journal also pertain to this production process. -

Symplekin and Multiple Other Polyadenylation Factors Participate in 3 -End Maturation of Histone Mrnas
Downloaded from genesdev.cshlp.org on September 27, 2021 - Published by Cold Spring Harbor Laboratory Press Symplekin and multiple other polyadenylation factors participate in 3-end maturation of histone mRNAs Nikolay G. Kolev and Joan A. Steitz1 Howard Hughes Medical Institute, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06536, USA .Most metazoan messenger RNAs encoding histones are cleaved, but not polyadenylated at their 3 ends Processing in mammalian cell extracts requires the U7 small nuclear ribonucleoprotein (U7 snRNP) and an unidentified heat-labile factor (HLF). We describe the identification of a heat-sensitive protein complex whose integrity is required for histone pre-mRNA cleavage. It includes all five subunits of the cleavage and polyadenylation specificity factor (CPSF), two subunits of the cleavage stimulation factor (CstF), and symplekin. Reconstitution experiments reveal that symplekin, previously shown to be necessary for cytoplasmic poly(A) tail elongation and translational activation of mRNAs during Xenopus oocyte maturation, is the essential heat-labile component. Thus, a common molecular machinery contributes to the nuclear maturation of mRNAs both lacking and possessing poly(A), as well as to cytoplasmic poly(A) tail elongation. [Keywords: Symplekin; polyadenylation; 3Ј-end processing; U7 snRNP; histone mRNA; Cajal body] Received September 1, 2005; revised version accepted September 12, 2005. During the S phase of the cell cycle, histone mRNA lev- unique component of the U7-specific Sm core, in which els are up-regulated to meet the need for histones to the spliceosomal SmD1 and SmD2 proteins are replaced package newly synthesized DNA. Increased transcrip- by Lsm10 and Lsm11 (Pillai et al. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Novel LRPPRC Compound Heterozygous Mutation in a Child
Piro et al. Italian Journal of Pediatrics (2020) 46:140 https://doi.org/10.1186/s13052-020-00903-7 CASE REPORT Open Access Novel LRPPRC compound heterozygous mutation in a child with early-onset Leigh syndrome French-Canadian type: case report of an Italian patient Ettore Piro1* , Gregorio Serra1, Vincenzo Antona1, Mario Giuffrè1, Elisa Giorgio2, Fabio Sirchia3, Ingrid Anne Mandy Schierz1, Alfredo Brusco2 and Giovanni Corsello1 Abstract Background: Mitochondrial diseases, also known as oxidative phosphorylation (OXPHOS) disorders, with a prevalence rate of 1:5000, are the most frequent inherited metabolic diseases. Leigh Syndrome French Canadian type (LSFC), is caused by mutations in the nuclear gene (2p16) leucine-rich pentatricopeptide repeat-containing (LRPPRC). It is an autosomal recessive neurogenetic OXPHOS disorder, phenotypically distinct from other types of Leigh syndrome, with a carrier frequency up to 1:23 and an incidence of 1:2063 in the Saguenay-Lac-St Jean region of Quebec. Recently, LSFC has also been reported outside the French-Canadian population. Patient presentation: We report a male Italian (Sicilian) child, born preterm at 28 + 6/7 weeks gestation, carrying a novel LRPPRC compound heterozygous mutation, with facial dysmorphisms, neonatal hypotonia, non-epileptic paroxysmal motor phenomena, and absent sucking-swallowing-breathing coordination requiring, at 4.5 months, a percutaneous endoscopic gastrostomy tube placement. At 5 months brain Magnetic Resonance Imagingshoweddiffusecortical atrophy, hypoplasia of corpus callosum, cerebellar vermis hypoplasia, and unfolded hippocampi. Both auditory and visual evoked potentials were pathological. In the following months Video EEG confirmed the persistence of sporadic non epileptic motor phenomena. No episode of metabolic decompensation, acidosis or ketosis, frequently observed in LSFC has been reported. -

Download Validation Data
PrimePCR™Assay Validation Report Gene Information Gene Name FAST kinase domain-containing protein 1 Gene Symbol Fastkd1 Organism Rat Gene Summary Description Not Available Gene Aliases Not Available RefSeq Accession No. NM_001191738 UniGene ID Rn.226110 Ensembl Gene ID ENSRNOG00000024335 Entrez Gene ID 311112 Assay Information Unique Assay ID qRnoCEP0034063 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENSRNOT00000036585 Amplicon Context Sequence AAAAAAAAAAACTACAGTCATGATCTGCCTGCTCCAAATATCTGTTCTCTCAGGTA GTCCATCCGTGTATCCTTCGTTGACATTGCCATGGAGTTCCA Amplicon Length (bp) 68 Chromosome Location 3:62537455-62537552 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 97 R2 0.9997 cDNA Cq 23.19 cDNA Tm (Celsius) 79 gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Fastkd1, Rat Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. Details for each transcript can be found on the Ensembl website at www.ensembl.org. -

A SARS-Cov-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing
A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing Supplementary Information Supplementary Discussion All SARS-CoV-2 protein and gene functions described in the subnetwork appendices, including the text below and the text found in the individual bait subnetworks, are based on the functions of homologous genes from other coronavirus species. These are mainly from SARS-CoV and MERS-CoV, but when available and applicable other related viruses were used to provide insight into function. The SARS-CoV-2 proteins and genes listed here were designed and researched based on the gene alignments provided by Chan et. al. 1 2020 . Though we are reasonably sure the genes here are well annotated, we want to note that not every protein has been verified to be expressed or functional during SARS-CoV-2 infections, either in vitro or in vivo. In an effort to be as comprehensive and transparent as possible, we are reporting the sub-networks of these functionally unverified proteins along with the other SARS-CoV-2 proteins. In such cases, we have made notes within the text below, and on the corresponding subnetwork figures, and would advise that more caution be taken when examining these proteins and their molecular interactions. Due to practical limits in our sample preparation and data collection process, we were unable to generate data for proteins corresponding to Nsp3, Orf7b, and Nsp16. Therefore these three genes have been left out of the following literature review of the SARS-CoV-2 proteins and the protein-protein interactions (PPIs) identified in this study. -

Building the Vertebrate Codex Using the Gene Breaking Protein Trap Library
Genetics, Development and Cell Biology Publications Genetics, Development and Cell Biology 2020 Building the vertebrate codex using the gene breaking protein trap library Noriko Ichino Mayo Clinic Gaurav K. Varshney National Institutes of Health Ying Wang Iowa State University Hsin-kai Liao Iowa State University Maura McGrail Iowa State University, [email protected] See next page for additional authors Follow this and additional works at: https://lib.dr.iastate.edu/gdcb_las_pubs Part of the Molecular Genetics Commons, and the Research Methods in Life Sciences Commons The complete bibliographic information for this item can be found at https://lib.dr.iastate.edu/ gdcb_las_pubs/261. For information on how to cite this item, please visit http://lib.dr.iastate.edu/howtocite.html. This Article is brought to you for free and open access by the Genetics, Development and Cell Biology at Iowa State University Digital Repository. It has been accepted for inclusion in Genetics, Development and Cell Biology Publications by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Building the vertebrate codex using the gene breaking protein trap library Abstract One key bottleneck in understanding the human genome is the relative under-characterization of 90% of protein coding regions. We report a collection of 1200 transgenic zebrafish strains made with the gene- break transposon (GBT) protein trap to simultaneously report and reversibly knockdown the tagged genes. Protein trap-associated mRFP expression shows previously undocumented expression of 35% and 90% of cloned genes at 2 and 4 days post-fertilization, respectively. Further, investigated alleles regularly show 99% gene-specific mRNA knockdown. -
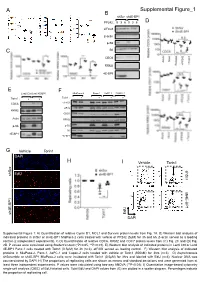
H Supplemental Figure 1 I D E B a C
A Supplemental Figure_1 B shScr sh4E-BP1 PP242 : 0 3 6 0 3 6 D eIF4G1 β-actin p-S6 C S6 CDC6 RRM2 4E-BP1 E F Lenti Ctrl Lenti 4E-BP1 MiaPaca-2 Panc-1 AsPC-1 Capan-2 Torin1 : Torin1 : + + + + + + CDC6 eIF4G1 eIF3a RRM2 CDC6 Actin RRM2 p-S6 p-S6 S6 4E-BP1 4E-BP1 G Vehicle Torin1 DAPI H I Vehicle Torin1 62.20% 45.30% shScr sh4E-BP1 EdU shScr 59.58% 49.47% EdU sh4E-BP1 DAPI Supplemental Figure 1: A) Quantification of relative Cyclin D1, MCL1 and Survivin protein levels from Fig. 1A. B) Western blot analysis of indicated proteins in shScr or sh4E-BP1 MiaPaca-2 cells treated with vehicle or PP242 (5µM) for 3h and 6h. β-actin served as a loading control (2 independent experiments). C-D) Quantification of relative CDC6, RRM2 and CDC7 protein levels from (C) Fig. 2C and (D) Fig. 2D. P values were calculated using Student’s t-test (*P<0.05, **P<0.01). E) Western blot analysis of indicated proteins in Lenti Ctrl or Lenti 4E-BP1 Panc-1 cells treated with Torin1 (0.5µM) for 3h (n=3). eIF4GI served as loading control. F) Western blot analysis of indicated proteins in MiaPaca-2, Panc-1, AsPC-1 and Capan-2 cells treated with vehicle or Torin1 (500nM) for 3hrs (n=3). G) Asynchronous shScramble or sh4E-BP1 MiaPaca-2 cells were incubated with Torin1 (0.5μM) for 3hrs and labeled with EdU (n=3). Nuclear DNA was counterstained by DAPI. H) The proportions of replicating cells are shown as means and standard deviations and were generated from at least three independent experiments. -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas.