Genelc Regulalon of Gene Expression Varialon
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Sialic Acids and Their Influence on Human NK Cell Function
cells Review Sialic Acids and Their Influence on Human NK Cell Function Philip Rosenstock * and Thomas Kaufmann Institute for Physiological Chemistry, Martin-Luther-University Halle-Wittenberg, Hollystr. 1, D-06114 Halle/Saale, Germany; [email protected] * Correspondence: [email protected] Abstract: Sialic acids are sugars with a nine-carbon backbone, present on the surface of all cells in humans, including immune cells and their target cells, with various functions. Natural Killer (NK) cells are cells of the innate immune system, capable of killing virus-infected and tumor cells. Sialic acids can influence the interaction of NK cells with potential targets in several ways. Different NK cell receptors can bind sialic acids, leading to NK cell inhibition or activation. Moreover, NK cells have sialic acids on their surface, which can regulate receptor abundance and activity. This review is focused on how sialic acids on NK cells and their target cells are involved in NK cell function. Keywords: sialic acids; sialylation; NK cells; Siglecs; NCAM; CD56; sialyltransferases; NKp44; Nkp46; NKG2D 1. Introduction 1.1. Sialic Acids N-Acetylneuraminic acid (Neu5Ac) is the most common sialic acid in the human organism and also the precursor for all other sialic acid derivatives. The biosynthesis of Neu5Ac begins in the cytosol with uridine diphosphate-N-acetylglucosamine (UDP- Citation: Rosenstock, P.; Kaufmann, GlcNAc) as its starting component [1]. It is important to understand that sialic acid T. Sialic Acids and Their Influence on formation is strongly linked to glycolysis, since it results in the production of fructose-6- Human NK Cell Function. Cells 2021, phosphate (F6P) and phosphoenolpyruvate (PEP). -

Human Lectins, Their Carbohydrate Affinities and Where to Find Them
biomolecules Review Human Lectins, Their Carbohydrate Affinities and Where to Review HumanFind Them Lectins, Their Carbohydrate Affinities and Where to FindCláudia ThemD. Raposo 1,*, André B. Canelas 2 and M. Teresa Barros 1 1, 2 1 Cláudia D. Raposo * , Andr1 é LAQVB. Canelas‐Requimte,and Department M. Teresa of Chemistry, Barros NOVA School of Science and Technology, Universidade NOVA de Lisboa, 2829‐516 Caparica, Portugal; [email protected] 12 GlanbiaLAQV-Requimte,‐AgriChemWhey, Department Lisheen of Chemistry, Mine, Killoran, NOVA Moyne, School E41 of ScienceR622 Co. and Tipperary, Technology, Ireland; canelas‐ [email protected] NOVA de Lisboa, 2829-516 Caparica, Portugal; [email protected] 2* Correspondence:Glanbia-AgriChemWhey, [email protected]; Lisheen Mine, Tel.: Killoran, +351‐212948550 Moyne, E41 R622 Tipperary, Ireland; [email protected] * Correspondence: [email protected]; Tel.: +351-212948550 Abstract: Lectins are a class of proteins responsible for several biological roles such as cell‐cell in‐ Abstract:teractions,Lectins signaling are pathways, a class of and proteins several responsible innate immune for several responses biological against roles pathogens. such as Since cell-cell lec‐ interactions,tins are able signalingto bind to pathways, carbohydrates, and several they can innate be a immuneviable target responses for targeted against drug pathogens. delivery Since sys‐ lectinstems. In are fact, able several to bind lectins to carbohydrates, were approved they by canFood be and a viable Drug targetAdministration for targeted for drugthat purpose. delivery systems.Information In fact, about several specific lectins carbohydrate were approved recognition by Food by andlectin Drug receptors Administration was gathered for that herein, purpose. plus Informationthe specific organs about specific where those carbohydrate lectins can recognition be found by within lectin the receptors human was body. -
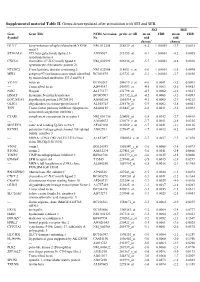
Supplemental Material Table II. Genes Downregulated After Stimulation with SEI and SEB
Supplemental material Table II. Genes downregulated after stimulation with SEI and SEB. SEI SEB Gene Gene Title NCBI Accession probe set ID mean FDR mean FDR Symbola No fold fold changeb changeb HEY1c hairy/enhancer-of-split related with YRPW NM_012258 218839_at -6.2 < 0.0001 -3.9 0.0015 motif 1 ST3GAL6c ST3 beta-galactoside alpha-2,3- AI989567 213355_at -6.1 < 0.0001 -4.2 0.0005 sialyltransferase 6 CXCL6 chemokine (C-X-C motif) ligand 6 NM_002993 206336_at -5.7 < 0.0001 -4.8 0.0010 (granulocyte chemotactic protein 2) NT5DC2 5'-nucleotidase domain containing 2 NM_022908 218051_s_at -5.6 < 0.0001 -2.6 0.0490 MFI2 antigen p97 (melanoma associated) identified BC001875 223723_at -5.3 < 0.0001 -3.7 0.0036 by monoclonal antibodies 133.2 and 96.5 VCANc versican BF590263 204619_s_at -4.6 0.0001 -5.2 0.0003 --- Transcribed locus AI494347 240393_at -4.5 0.0003 -2.6 0.0413 NOG Noggin AL575177 231798_at -4.5 0.0002 -2.6 0.0413 HNMTc histamine N-methyltransferase BC005907 211732_x_at -4.2 0.0006 -3.2 0.0093 LOC285181 hypothetical protein LOC285181 AA002166 1561334_at -4.1 0.0005 -2.9 0.0212 OLIG1 oligodendrocyte transcription factor 1 AL355743 228170_at -3.9 0.0002 -3.8 0.0021 TFPI Tissue factor pathway inhibitor (lipoprotein- AL080215 215447_at -3.8 0.0011 -3.2 0.0092 associated coagulation inhibitor) C5AR1 complement component 5a receptor 1 NM_001736 220088_at -3.8 0.0012 -2.7 0.0418 --- --- AA805633 230175_s_at -3.7 0.0011 -2.8 0.0326 SIGLEC9 sialic acid binding Ig-like lectin 9 AF247180 210569_s_at -3.7 0.0011 -3.1 0.0136 KCNE3 potassium voltage-gated -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

1 SUPPLEMENTAL DATA Figure S1. Poly I:C Induces IFN-Β Expression
SUPPLEMENTAL DATA Figure S1. Poly I:C induces IFN-β expression and signaling. Fibroblasts were incubated in media with or without Poly I:C for 24 h. RNA was isolated and processed for microarray analysis. Genes showing >2-fold up- or down-regulation compared to control fibroblasts were analyzed using Ingenuity Pathway Analysis Software (Red color, up-regulation; Green color, down-regulation). The transcripts with known gene identifiers (HUGO gene symbols) were entered into the Ingenuity Pathways Knowledge Base IPA 4.0. Each gene identifier mapped in the Ingenuity Pathways Knowledge Base was termed as a focus gene, which was overlaid into a global molecular network established from the information in the Ingenuity Pathways Knowledge Base. Each network contained a maximum of 35 focus genes. 1 Figure S2. The overlap of genes regulated by Poly I:C and by IFN. Bioinformatics analysis was conducted to generate a list of 2003 genes showing >2 fold up or down- regulation in fibroblasts treated with Poly I:C for 24 h. The overlap of this gene set with the 117 skin gene IFN Core Signature comprised of datasets of skin cells stimulated by IFN (Wong et al, 2012) was generated using Microsoft Excel. 2 Symbol Description polyIC 24h IFN 24h CXCL10 chemokine (C-X-C motif) ligand 10 129 7.14 CCL5 chemokine (C-C motif) ligand 5 118 1.12 CCL5 chemokine (C-C motif) ligand 5 115 1.01 OASL 2'-5'-oligoadenylate synthetase-like 83.3 9.52 CCL8 chemokine (C-C motif) ligand 8 78.5 3.25 IDO1 indoleamine 2,3-dioxygenase 1 76.3 3.5 IFI27 interferon, alpha-inducible -
Reviewed by HLDA1
Human CD Marker Chart Reviewed by HLDA1 T Cell Key Markers CD3 CD4 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD8 CD1a R4, T6, Leu6, HTA1 b-2-Microglobulin, CD74 + + + – + – – – CD74 DHLAG, HLADG, Ia-g, li, invariant chain HLA-DR, CD44 + + + + + + CD158g KIR2DS5 + + CD248 TEM1, Endosialin, CD164L1, MGC119478, MGC119479 Collagen I/IV Fibronectin + ST6GAL1, MGC48859, SIAT1, ST6GALL, ST6N, ST6 b-Galactosamide a-2,6-sialyl- CD1b R1, T6m Leu6 b-2-Microglobulin + + + – + – – – CD75 CD22 CD158h KIR2DS1, p50.1 HLA-C + + CD249 APA, gp160, EAP, ENPEP + + tranferase, Sialo-masked lactosamine, Carbohydrate of a2,6 sialyltransferase + – – + – – + – – CD1c M241, R7, T6, Leu6, BDCA1 b-2-Microglobulin + + + – + – – – CD75S a2,6 Sialylated lactosamine CD22 (proposed) + + – – + + – + + + CD158i KIR2DS4, p50.3 HLA-C + – + CD252 TNFSF4, -

UNIVERSITY of CALIFORNIA Los Angeles Disease
UNIVERSITY OF CALIFORNIA Los Angeles Disease-specific differences in glycosylation of mouse and human skeletal muscle A dissertation submitted in partial satisfaction of the requirements for the Degree of Philosophy in Cellular and Molecular Pathology by Brian James McMorran 2017 © Copyright by Brian James McMorran 2017 ABSTRACT OF THE DISSERTATION Disease-specific differences in glycosylation of mouse and human skeletal muscle by Brian James McMorran Doctor of Philosophy in Cellular and Molecular Pathology University of California, Los Angeles, 2017 Professor Linda G. Baum, Chair Proper glycosylation of proteins at the muscle cell membrane, or sarcolemma, is critical for proper muscle function. The laminin receptor alpha-dystroglycan (α-DG) is heavily glycosylated and mutations in 24 genes involved in proper α-DG glycosylation have been identified as causing various forms of congenital muscular dystrophy. While work over the past decade has elucidated the structure bound by laminin and the enzymes required for its creation, very little is known about muscle glycosylation outside of α-DG glycosylation. The modification of glycan structures with terminal GalNAc residues at the rodent neuromuscular junction (NMJ) has remained the focus of work in mouse muscle glycosylation, while qualitative lectin histochemistry studies performed three decades ago represent the majority of human muscle glycosylation research. This thesis quantifies differentiation-, species-, and disease-specific differences in mouse and human skeletal muscle glycosylation. Following differentiation of mouse myotubes, increased binding was found of lectins specific for GalNAc and O-glycans. Additionally, ii analysis of binding preferences of four GalNAc-specific lectins, which historically have been used to identify the rodent NMJ, identified differences in the glycan types bound on distinct glycoproteins by each lectin. -

Identification of Biomarkers, Pathways and Potential Therapeutic Targets for Heart Failure Using Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.05.455244; this version posted August 6, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Identification of biomarkers, pathways and potential therapeutic targets for heart failure using bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2021.08.05.455244; this version posted August 6, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Heart failure (HF) is a complex cardiovascular diseases associated with high mortality. To discover key molecular changes in HF, we analyzed next-generation sequencing (NGS) data of HF. In this investigation, differentially expressed genes (DEGs) were analyzed using limma in R package from GSE161472 of the Gene Expression Omnibus (GEO). Then, gene enrichment analysis, protein-protein interaction (PPI) network, miRNA-hub gene regulatory network and TF-hub gene regulatory network construction, and topological analysis were performed on the DEGs by the Gene Ontology (GO), REACTOME pathway, STRING, HiPPIE, miRNet, NetworkAnalyst and Cytoscape. Finally, we performed receiver operating characteristic curve (ROC) analysis of hub genes. A total of 930 DEGs 9464 up regulated genes and 466 down regulated genes) were identified in HF. -

Influence of SIGLEC9 Polymorphisms on COPD Phenotypes Including Exacerbation
1 Influence of SIGLEC9 polymorphisms on COPD phenotypes including exacerbation 2 frequency 3 4 Takeo Ishii, MD, PhD1,2, Takashi Angata, PhD3,4, Emily S Wan, MD, MPH5, 6, Michael H Cho, 5 MD, MPH5, 6, Takashi Motegi, MD, PhD1,2, Congxiao Gao MD, PhD4, Kazuaki Ohtsubo, PhD7, 6 Shinobu Kitazume, PhD4, Akihiko Gemma, MD, PhD2, Peter D. Pare, MD8, David A. Lomas, 7 MD, PhD9, Edwin K Silverman, MD, PhD5, 6, Naoyuki Taniguchi, MD, PhD4, and Kozui Kida, 8 MD, PhD1,2 9 10 1Respiratory Care Clinic, Nippon Medical School, 4-7-15-8F Kudan-Minami, Chiyoda, Tokyo 11 102-0074, Japan; 2Division of Pulmonary Medicine, Infectious Diseases and Oncology, 12 Department of Internal Medicine, Nippon Medical School, 1-1-5 Sendagi, Bunkyo, Tokyo 113- 13 8602, Japan; 3Institute of Biological Chemistry, Academia Sinica, 128 Section 2, Academia 14 Road, Nankang, Taipei 11529, Taiwan; 4Disease Glycomics Team, Systems Glycobiology 15 Research Group, Global Research Cluster, RIKEN, 2-1 Hirosawa, Wako, Saitama 351-0198, 16 Japan.; 5Channing Division of Network Medicine, Brigham and Women’s Hospital, 181 17 Longwood Avenue, Boston, MA 02115, USA.; 6Division of Pulmonary and Critical Care, 1 Brigham and Women’s Hospital, Boston, MA, USA; 7Department of Analytical biochemistry, 2 Faculty of Life Sciences, Kumamoto University 4-24-1 Kuhonji, Chuo, Kumamoto, 862-0976, 3 Japan ;8 University of British Columibia Center for Heart Lung Innovation, University of St 4 Paul’s Hospital, British Columbia, Canada; 9UCL Respiratory, University College London, 5 Rayne Building, London, United Kingdom. 6 7 Correspondence should be addressed to: 8 Kozui Kida, M.D., Respiratory Care Clinic, Nippon Medical School, 4-7-15 Kudan-Minami, 9 Chiyoda-Ku, Tokyo 102-0074, Japan 10 Tel.: +81-3-5276-2325; Fax: +81-3-5276-2326; Email: [email protected] 11 12 Summary at a Glance 13 A haplotype of SIGLEC9 gene was associated with exacerbation frequency and emphysema in 14 Japanese COPD patients (but not in ECLIPSE cohort). -

Secreted Ectodomain of Sialic Acid-Binding Ig-Like Lectin-9 And
2452 • The Journal of Neuroscience, February 11, 2015 • 35(6):2452–2464 Development/Plasticity/Repair Secreted Ectodomain of Sialic Acid-Binding Ig-Like Lectin-9 and Monocyte Chemoattractant Protein-1 Promote Recovery after Rat Spinal Cord Injury by Altering Macrophage Polarity Kohki Matsubara,1 XYoshihiro Matsushita,1 Kiyoshi Sakai,1 Fumiya Kano,1 Megumi Kondo,1 XMariko Noda,4 Noboru Hashimoto,3 Shiro Imagama,2 Naoki Ishiguro,2 Akio Suzumura,4 Minoru Ueda,1 Koichi Furukawa,3 and Akihito Yamamoto1 1Department of Oral and Maxillofacial Surgery, 2Orthopedic Surgery, and 3Biochemistry II, Nagoya University Graduate School of Medicine, Showa-ku, Nagoya 466-8550, Japan, and 4Department of Neuroimmunology, Research Institute of Environmental Medicine, Nagoya University, Chikusa-ku, Nagoya 464-8601, Japan Engrafted mesenchymal stem cells from human deciduous dental pulp (SHEDs) support recovery from neural insults via paracrine mechanisms that are poorly understood. Here we show that the conditioned serum-free medium (CM) from SHEDs, administered intrathecally into rat injured spinal cord during the acute postinjury period, caused remarkable functional recovery. The ability of SHED-CM to induce recovery was associated with an immunoregulatory activity that induced anti-inflammatory M2-like macrophages. Secretome analysis of the SHED-CM revealed a previously unrecognized set of inducers for anti-inflammatory M2-like macrophages: monocytechemoattractantprotein-1(MCP-1)andthesecretedectodomainofsialicacid-bindingIg-likelectin-9(ED-Siglec-9).Depleting MCP-1 and ED-Siglec-9 from the SHED-CM prominently reduced its ability to induce M2-like macrophages and to promote functional recovery after spinal cord injury (SCI). The combination of MCP-1 and ED-Siglec-9 synergistically promoted the M2-like differentiation of bone marrow-derived macrophages in vitro, and this effect was abolished by a selective antagonist for CC chemokine receptor 2 (CCR2) or by the genetic knock-out of CCR2.