Investigation of Siglec8 As a Therapeutic Target to Treat Asthma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mast Cells Promote Seasonal White Adipose Beiging in Humans
Diabetes Volume 66, May 2017 1237 Mast Cells Promote Seasonal White Adipose Beiging in Humans Brian S. Finlin,1 Beibei Zhu,1 Amy L. Confides,2 Philip M. Westgate,3 Brianna D. Harfmann,1 Esther E. Dupont-Versteegden,2 and Philip A. Kern1 Diabetes 2017;66:1237–1246 | DOI: 10.2337/db16-1057 Human subcutaneous (SC) white adipose tissue (WAT) localized to the neck and thorax of humans (4–8), and in a increases the expression of beige adipocyte genes in the process known as beiging (9), UCP1-positive adipocytes winter. Studies in rodents suggest that a number of form in subcutaneous (SC) white adipose tissue (WAT) immune mediators are important in the beiging response. (10). Beige adipocytes have unique developmental origins, We studied the seasonal beiging response in SC WAT gene signatures, and functional properties, including being from lean humans. We measured the gene expression of highly inducible to increase UCP1 in response to catechol- various immune cell markers and performed multivariate amines (9,11–13). Although questions exist about whether analysis of the gene expression data to identify genes beige fat can make a meaningful contribution to energy OBESITY STUDIES that predict UCP1. Interleukin (IL)-4 and, unexpectedly, expenditure in humans (reviewed in Porter et al. [14]), the mast cell marker CPA3 predicted UCP1 gene expres- the induction of beige fat in rodent models is associated sion. Therefore, we investigated the effects of mast with increased energy expenditure and improved glucose cells on UCP1 induction by adipocytes. TIB64 mast cells homeostasis (13). responded to cold by releasing histamine and IL-4, and this medium stimulated UCP1 expression and lipolysis by Activation of the sympathetic nervous system by cold 3T3-L1 adipocytes. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Sialic Acids and Their Influence on Human NK Cell Function
cells Review Sialic Acids and Their Influence on Human NK Cell Function Philip Rosenstock * and Thomas Kaufmann Institute for Physiological Chemistry, Martin-Luther-University Halle-Wittenberg, Hollystr. 1, D-06114 Halle/Saale, Germany; [email protected] * Correspondence: [email protected] Abstract: Sialic acids are sugars with a nine-carbon backbone, present on the surface of all cells in humans, including immune cells and their target cells, with various functions. Natural Killer (NK) cells are cells of the innate immune system, capable of killing virus-infected and tumor cells. Sialic acids can influence the interaction of NK cells with potential targets in several ways. Different NK cell receptors can bind sialic acids, leading to NK cell inhibition or activation. Moreover, NK cells have sialic acids on their surface, which can regulate receptor abundance and activity. This review is focused on how sialic acids on NK cells and their target cells are involved in NK cell function. Keywords: sialic acids; sialylation; NK cells; Siglecs; NCAM; CD56; sialyltransferases; NKp44; Nkp46; NKG2D 1. Introduction 1.1. Sialic Acids N-Acetylneuraminic acid (Neu5Ac) is the most common sialic acid in the human organism and also the precursor for all other sialic acid derivatives. The biosynthesis of Neu5Ac begins in the cytosol with uridine diphosphate-N-acetylglucosamine (UDP- Citation: Rosenstock, P.; Kaufmann, GlcNAc) as its starting component [1]. It is important to understand that sialic acid T. Sialic Acids and Their Influence on formation is strongly linked to glycolysis, since it results in the production of fructose-6- Human NK Cell Function. Cells 2021, phosphate (F6P) and phosphoenolpyruvate (PEP). -

Human Lectins, Their Carbohydrate Affinities and Where to Find Them
biomolecules Review Human Lectins, Their Carbohydrate Affinities and Where to Review HumanFind Them Lectins, Their Carbohydrate Affinities and Where to FindCláudia ThemD. Raposo 1,*, André B. Canelas 2 and M. Teresa Barros 1 1, 2 1 Cláudia D. Raposo * , Andr1 é LAQVB. Canelas‐Requimte,and Department M. Teresa of Chemistry, Barros NOVA School of Science and Technology, Universidade NOVA de Lisboa, 2829‐516 Caparica, Portugal; [email protected] 12 GlanbiaLAQV-Requimte,‐AgriChemWhey, Department Lisheen of Chemistry, Mine, Killoran, NOVA Moyne, School E41 of ScienceR622 Co. and Tipperary, Technology, Ireland; canelas‐ [email protected] NOVA de Lisboa, 2829-516 Caparica, Portugal; [email protected] 2* Correspondence:Glanbia-AgriChemWhey, [email protected]; Lisheen Mine, Tel.: Killoran, +351‐212948550 Moyne, E41 R622 Tipperary, Ireland; [email protected] * Correspondence: [email protected]; Tel.: +351-212948550 Abstract: Lectins are a class of proteins responsible for several biological roles such as cell‐cell in‐ Abstract:teractions,Lectins signaling are pathways, a class of and proteins several responsible innate immune for several responses biological against roles pathogens. such as Since cell-cell lec‐ interactions,tins are able signalingto bind to pathways, carbohydrates, and several they can innate be a immuneviable target responses for targeted against drug pathogens. delivery Since sys‐ lectinstems. In are fact, able several to bind lectins to carbohydrates, were approved they by canFood be and a viable Drug targetAdministration for targeted for drugthat purpose. delivery systems.Information In fact, about several specific lectins carbohydrate were approved recognition by Food by andlectin Drug receptors Administration was gathered for that herein, purpose. plus Informationthe specific organs about specific where those carbohydrate lectins can recognition be found by within lectin the receptors human was body. -
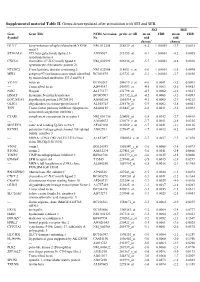
Supplemental Material Table II. Genes Downregulated After Stimulation with SEI and SEB
Supplemental material Table II. Genes downregulated after stimulation with SEI and SEB. SEI SEB Gene Gene Title NCBI Accession probe set ID mean FDR mean FDR Symbola No fold fold changeb changeb HEY1c hairy/enhancer-of-split related with YRPW NM_012258 218839_at -6.2 < 0.0001 -3.9 0.0015 motif 1 ST3GAL6c ST3 beta-galactoside alpha-2,3- AI989567 213355_at -6.1 < 0.0001 -4.2 0.0005 sialyltransferase 6 CXCL6 chemokine (C-X-C motif) ligand 6 NM_002993 206336_at -5.7 < 0.0001 -4.8 0.0010 (granulocyte chemotactic protein 2) NT5DC2 5'-nucleotidase domain containing 2 NM_022908 218051_s_at -5.6 < 0.0001 -2.6 0.0490 MFI2 antigen p97 (melanoma associated) identified BC001875 223723_at -5.3 < 0.0001 -3.7 0.0036 by monoclonal antibodies 133.2 and 96.5 VCANc versican BF590263 204619_s_at -4.6 0.0001 -5.2 0.0003 --- Transcribed locus AI494347 240393_at -4.5 0.0003 -2.6 0.0413 NOG Noggin AL575177 231798_at -4.5 0.0002 -2.6 0.0413 HNMTc histamine N-methyltransferase BC005907 211732_x_at -4.2 0.0006 -3.2 0.0093 LOC285181 hypothetical protein LOC285181 AA002166 1561334_at -4.1 0.0005 -2.9 0.0212 OLIG1 oligodendrocyte transcription factor 1 AL355743 228170_at -3.9 0.0002 -3.8 0.0021 TFPI Tissue factor pathway inhibitor (lipoprotein- AL080215 215447_at -3.8 0.0011 -3.2 0.0092 associated coagulation inhibitor) C5AR1 complement component 5a receptor 1 NM_001736 220088_at -3.8 0.0012 -2.7 0.0418 --- --- AA805633 230175_s_at -3.7 0.0011 -2.8 0.0326 SIGLEC9 sialic acid binding Ig-like lectin 9 AF247180 210569_s_at -3.7 0.0011 -3.1 0.0136 KCNE3 potassium voltage-gated -

Pancancer IO360 Human Vapril2018
Gene Name Official Full Gene name Alias/Prev Symbols Previous Name(s) Alias Symbol(s) Alias Name(s) A2M alpha-2-macroglobulin FWP007,S863-7,CPAMD5 ABCF1 ATP binding cassette subfamily F member 1 ABC50 ATP-binding cassette, sub-family F (GCN20),EST123147 member 1 ACVR1C activin A receptor type 1C activin A receptor, type IC ALK7,ACVRLK7 ADAM12 ADAM metallopeptidase domain 12 a disintegrin and metalloproteinase domainMCMPMltna,MLTN 12 (meltrin alpha) meltrin alpha ADGRE1 adhesion G protein-coupled receptor E1 TM7LN3,EMR1 egf-like module containing, mucin-like, hormone receptor-like sequence 1,egf-like module containing, mucin-like, hormone receptor-like 1 ADM adrenomedullin AM ADORA2A adenosine A2a receptor ADORA2 RDC8 AKT1 AKT serine/threonine kinase 1 v-akt murine thymoma viral oncogene homologRAC,PKB,PRKBA,AKT 1 ALDOA aldolase, fructose-bisphosphate A aldolase A, fructose-bisphosphate ALDOC aldolase, fructose-bisphosphate C aldolase C, fructose-bisphosphate ANGPT1 angiopoietin 1 KIAA0003,Ang1 ANGPT2 angiopoietin 2 Ang2 ANGPTL4 angiopoietin like 4 angiopoietin-like 4 pp1158,PGAR,ARP4,HFARP,FIAF,NL2fasting-induced adipose factor,hepatic angiopoietin-related protein,PPARG angiopoietin related protein,hepatic fibrinogen/angiopoietin-related protein,peroxisome proliferator-activated receptor (PPAR) gamma induced angiopoietin-related protein,angiopoietin-related protein 4 ANLN anillin actin binding protein anillin (Drosophila Scraps homolog), actin bindingANILLIN,Scraps,scra protein,anillin, actin binding protein (scraps homolog, Drosophila) -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

Suppressive Myeloid Cells Are a Hallmark of Severe COVID-19
medRxiv preprint doi: https://doi.org/10.1101/2020.06.03.20119818; this version posted June 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . 1 Suppressive myeloid cells are a hallmark of 2 severe COVID-19 3 Jonas Schulte-Schrepping1*, Nico Reusch1*, Daniela Paclik2*, Kevin Baßler1*, Stephan 4 Schlickeiser3*, Bowen Zhang4*, Benjamin Krämer5*, Tobias Krammer6*, Sophia Brumhard7*, 5 Lorenzo Bonaguro1*, Elena De Domenico8*, Daniel Wendisch7*, Martin Grasshoff4, Theodore S. 6 Kapellos1, Michael Beckstette4, Tal Pecht1, Adem Saglam8, Oliver Dietrich6, Henrik E. Mei9, Axel 7 R. Schulz9, Claudia Conrad7, Désirée Kunkel10, Ehsan Vafadarnejad6, Cheng-Jian Xu4,11, Arik 8 Horne1, Miriam Herbert1, Anna Drews8, Charlotte Thibeault7, Moritz Pfeiffer7, Stefan 9 Hippenstiel7,12, Andreas Hocke7,12, Holger Müller-Redetzky7, Katrin-Moira Heim7, Felix Machleidt7, 10 Alexander Uhrig7, Laure Bousquillon de Jarcy7, Linda Jürgens7, Miriam Stegemann7, Christoph 11 R. Glösenkamp7, Hans-Dieter Volk2,3,13, Christine Goffinet14,15, Jan Raabe5, Kim Melanie Kaiser5, 12 Michael To Vinh5, Gereon Rieke5, Christian Meisel14, Thomas Ulas8, Matthias Becker8, Robert 13 Geffers16, Martin Witzenrath7,12, Christian Drosten14,19, Norbert Suttorp7,12, Christof von Kalle17, 14 Florian Kurth7,18, Kristian Händler8, Joachim L. Schultze1,8,#,$, Anna C Aschenbrenner20,#, Yang 15 Li4,#, -

Impact of High Altitude Therapy on Type‐2 Immune Responses In
DR. TADECH BOONPIYATHAD (Orcid ID : 0000-0001-8690-7647) DR. GE TAN (Orcid ID : 0000-0003-0026-8739) MRS. MÜBECCEL AKDIS (Orcid ID : 0000-0002-9228-4594) Article type : Original Article: Basic and Translational Allergy Immunology Impact of High Altitude Therapy on Type-2 Immune Responses in Asthma Patients Tadech Boonpiyathad1,2,3, Gertruda Capova3,4, Hans-Werner Duchna3,4, Andrew L. Croxford5, Herve Farine5, Anita Dreher1,4, Martine Clozel5, Juliane Schreiber4, Petr Kubena4, Nonhlanhla Lunjani1,3, David Mirer1, Beate Rückert1, Pattraporn 1,3 1 5 1 Article Satitsuksanoa , Ge Tan , Peter M.A. Groenen , Mübeccel Akdis , Daniel S. Strasser5, Ellen D. Renner3,4,6, Cezmi A. Akdis1,3 1Swiss Institute of Allergy and Asthma Research, University of Zurich, Davos, Switzerland 2Department of Medicine, Phramongkutklao Hospital, Bangkok, Thailand 3Christine Kühne Center for Allergy Research and Education, Davos, Switzerland 4Hochgebirgsklinik Davos, Davos, Switzerland 5Drug Discovery, Idorsia Pharmaceuticals Ltd., Allschwil, Switzerland 6Chair and Institute of Enviromental Medicine – UNIKA-T, TU Munich and Helmholtz Zentrum Munich, Germany Corresponding author: Cezmi A. Akdis Swiss Institute of Allergy and Asthma Research, Obere Strasse 22, CH-7270, Davos Platz, Switzerland Email: [email protected] Tel.: +41 81 140 0848 Fax: +41 81 410 0840 This article has been accepted for publication and undergone full peer review but has not Accepted been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1111/all.13967 This article is protected by copyright. All rights reserved. Short title Immune response of asthma treatment in high altitude Abstract Background: Asthma patients present with distinct immunological profiles, with a predominance of type 2 endotype. -

Curriculum Vitae
Th17 Cells in Gingival Immunity and Their Key Role in Periodontitis Pathogenesis Item Type dissertation Authors Dutzan, Nicolas Publication Date 2017 Abstract Periodontitis is a very common human disease characterized by inflammatory bone destruction in the oral cavity. It affects more than 64 million adults in the United States and is often linked to systemic or distant co-morbidities. T helper (Th) cell... Keywords bone loss; IL-17; STAT3; Interleukin-17; Periodontitis; Th17 Cells Download date 27/09/2021 19:02:00 Link to Item http://hdl.handle.net/10713/7340 Curriculum Vitae Name: Nicolas Raul Dutzan Munoz E-mail: [email protected] Education: 2003 DDS, University of Chile, Chile 2008 MSc in Periodontology, University of Chile, Chile 2011 Certificate in Periodontics, University of Chile, Chile 2017 PhD, Oral and Experimental Pathology, Graduate School, University of Maryland Baltimore/National Institutes of Health Brief Chronology of Employment: 2003-2013 Private Dental Practice, Santiago, Chile. 2003-2013 Research Fellow, Periodontal Biology Laboratory, Conservative Dentistry Department, Faculty of Dentistry, University of Chile, Chile. 2007-Today Assistant Professor, Conservative Dentistry Department, Faculty of Dentistry, University of Chile, Chile. 2013-Today Visiting Fellow, National Institute of Dental and Craniofacial Research, National Institutes of Health. Professional Society Membership: International Association for Dental Research Sociedad Chilena de Periodoncia American Association of Immunologists Society for Mucosal Immunology Asociación Chilena de Inmunología Academic Honors and Awards: 2005 International Association for Dental Research, Division Chile, Distinguished Annual Research Award. 2005 Dentistry Faculty Scholarship for the MSc in Periodontology, University of Chile. 2005 Dentistry Faculty Scholarship for the Periodontics Residency, University of Chile. -

Revealing the Acute Asthma Ignorome: Characterization and Validation of Uninvestigated Gene Networks
www.nature.com/scientificreports OPEN Revealing the acute asthma ignorome: characterization and validation of uninvestigated gene Received: 07 December 2015 Accepted: 01 April 2016 networks Published: 21 April 2016 Michela Riba1,*, Jose Manuel Garcia Manteiga1,*, Berislav Bošnjak2,*, Davide Cittaro1, Pavol Mikolka2,†, Connie Le2,‡, Michelle M. Epstein2,# & Elia Stupka1,# Systems biology provides opportunities to fully understand the genes and pathways in disease pathogenesis. We used literature knowledge and unbiased multiple data meta-analysis paradigms to analyze microarray datasets across different mouse strains and acute allergic asthma models. Our combined gene-driven and pathway-driven strategies generated a stringent signature list totaling 933 genes with 41% (440) asthma-annotated genes and 59% (493) ignorome genes, not previously associated with asthma. Within the list, we identified inflammation, circadian rhythm, lung-specific insult response, stem cell proliferation domains, hubs, peripheral genes, and super-connectors that link the biological domains (Il6, Il1ß, Cd4, Cd44, Stat1, Traf6, Rela, Cadm1, Nr3c1, Prkcd, Vwf, Erbb2). In conclusion, this novel bioinformatics approach will be a powerful strategy for clinical and across species data analysis that allows for the validation of experimental models and might lead to the discovery of novel mechanistic insights in asthma. Allergen exposure causes a complex interaction of cellular and molecular networks leading to allergic asthma in susceptible individuals. Experimental mouse models of allergic asthma are widely used to understand disease pathogenesis and elucidate mechanisms underlying the initiation of allergic asthma1. For example, gene profiling of lung tissue from experimental mice during the initiation of allergic asthma in experiments with different proto- cols2–7 validated well-known genes and identified new genes with roles in disease pathogenesis such as C53, Arg18, Adam89, and Pon17, and dissected pathways activated by Il1310 and Stat611.