Parallel Evolution of Genes and Languages in the Caucasus Region
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ethnic Competition, Radical Islam and Challenges to Stability in the Republic of Dagestan
Ethnic Competition, Radical Islam, and Challenges to Stability in the Republic of Dagestan Edward C. Holland Doctoral Candidate Institute of Behavioral Science and Department of Geography Campus Box 487 University of Colorado Boulder, CO. 80309-0487 USA Email: [email protected] Phone: 1-303-492-4347; fax 1-303-492-3609 John O’Loughlin College Professor of Distinction and Professor of Geography Institute of Behavioral Science Campus Box 487 University of Colorado Boulder, CO. 80309-0487 USA Email: [email protected] Phone: 1-303-492-1619; fax 1-303-492-3609 Forthcoming COMMUNIST AND POST-COmMUNIST STUDIES 2010 Acknowledgements: This research was supported by a grant from the Human and Social Dynamics Initiative of the U.S. National Science Foundation, grant number 0433927. Thanks to Vladimir Kolossov, and Gearoid Ó Tuathail for help with the survey and the fieldwork in Dagestan, to our Dagestani colleagues, Eldar Eldarov, Shakhmardan Muduyev, Sharafudin Aliyev, and Zagir Atayev for their overwhelming hospitality on two field expeditions and to Nancy Thorwardson of the Institute of Behavioral Science for preparing the map for publication. We also acknowledge Alexei Grazhdankin and his colleagues of the Levada Center, Moscow for their timely, professional and friendly cooperation that ensured the success of the complex survey in Dagestan. Abstract Previous academic work on stability in Dagestan has focused on two potential cleavages, the republic’s ethnic diversity and the challenge from radical Islamist groups. Using results from a December 2005 survey, and focusing on Dagestan’s six main ethnic groups, this paper investigates attitudes towards the dual topics of the politicization of ethnicity and the relationship between terrorism and Islamism. -

The North Caucasus: the Challenges of Integration (III), Governance, Elections, Rule of Law
The North Caucasus: The Challenges of Integration (III), Governance, Elections, Rule of Law Europe Report N°226 | 6 September 2013 International Crisis Group Headquarters Avenue Louise 149 1050 Brussels, Belgium Tel: +32 2 502 90 38 Fax: +32 2 502 50 38 [email protected] Table of Contents Executive Summary ................................................................................................................... i Recommendations..................................................................................................................... iii I. Introduction ..................................................................................................................... 1 II. Russia between Decentralisation and the “Vertical of Power” ....................................... 3 A. Federative Relations Today ....................................................................................... 4 B. Local Government ...................................................................................................... 6 C. Funding and budgets ................................................................................................. 6 III. Elections ........................................................................................................................... 9 A. State Duma Elections 2011 ........................................................................................ 9 B. Presidential Elections 2012 ...................................................................................... -

Complete Bibliography (PDF)
COMPLETE BIBLIOGRAPHY OF THE PUBLICATIONS OF JOSEPH H. GREENBERG This bibliography follows the format of the bibliography published in On language (item 204, 723-37; with emendations) and the corrected supplemental bibliography in Language 78.560-64 (2002). The asterisked items have translations or reprints listed in the appendix to the bibliography. 1940 1. The decipherment of the Ben-Ali Diary, a preliminary statement. Journal of Negro History 25.372-75. 1941 *2. Some aspects of Negro-Mohammedan culture-contact among the Hausa. American Anthropologist 43.51-61. 3. Some problems in Hausa phonology. Language 17.316-23. 1946 *4. The influence of Islam on a Sudanese religion. New York: J.J. Augustin. Pp. ix, 73. 1947 5. Arabic loan-words in Hausa. Word 3.85-97. 6. Islam and clan organization among the Hausa. Southwestern Journal of Anthropology 3.193-211. *7. Swahili prosody. Journal of the American Oriental Society 67.24-30. 1948 8. The classification of African languages. American Anthropologist 50.24-30. *9. Linguistics and ethnology. Southwestern Journal of Anthropology 4.140-47. 10. The tonal system of Proto-Bantu. Word 4.196-208. 11. Review of H. Courlander & G. Herzog, The Cow-tail Switch and other West African tales. Journal of American Folklore 51.99-100. 1949 *12. Hausa verse prosody. Journal of the American Oriental Society 69.125-35. 13. The logical analysis of kinship. Philosophy of Science 16.58-64. *14. The Negro kingdoms of the Sudan. Transactions of the New York Academy of Sciences, Series II, 11.126-34. *15. Studies in African linguistic classification: I. -

An Amerind Etymological Dictionary
An Amerind Etymological Dictionary c 2007 by Merritt Ruhlen ! Printed in the United States of America Library of Congress Cataloging-in-Publication Data Greenberg, Joseph H. Ruhlen, Merritt An Amerind Etymological Dictionary Bibliography: p. Includes indexes. 1. Amerind Languages—Etymology—Classification. I. Title. P000.G0 2007 000!.012 00-00000 ISBN 0-0000-0000-0 (alk. paper) This book is dedicated to the Amerind people, the first Americans Preface The present volume is a revison, extension, and refinement of the ev- idence for the Amerind linguistic family that was initially offered in Greenberg (1987). This revision entails (1) the correction of a num- ber of forms, and the elimination of others, on the basis of criticism by specialists in various Amerind languages; (2) the consolidation of certain Amerind subgroup etymologies (given in Greenberg 1987) into Amerind etymologies; (3) the addition of many reconstructions from different levels of Amerind, based on a comprehensive database of all known reconstructions for Amerind subfamilies; and, finally, (4) the addition of a number of new Amerind etymologies presented here for the first time. I believe the present work represents an advance over the original, but it is at the same time simply one step forward on a project that will never be finished. M. R. September 2007 Contents Introduction 1 Dictionary 11 Maps 272 Classification of Amerind Languages 274 References 283 Semantic Index 296 Introduction This volume presents the lexical and grammatical evidence that defines the Amerind linguistic family. The evidence is presented in terms of 913 etymolo- gies, arranged alphabetically according to the English gloss. -

Russia's Peacetime Demographic Crisis
the national bureau of asian research nbr project report | may 2010 russia’s peacetime demographic crisis: Dimensions, Causes, Implications By Nicholas Eberstadt ++ The NBR Project Report provides access to current research on special topics conducted by the world’s leading experts in Asian affairs. The views expressed in these reports are those of the authors and do not necessarily reflect the views of other NBR research associates or institutions that support NBR. The National Bureau of Asian Research is a nonprofit, nonpartisan research institution dedicated to informing and strengthening policy. NBR conducts advanced independent research on strategic, political, economic, globalization, health, and energy issues affecting U.S. relations with Asia. Drawing upon an extensive network of the world’s leading specialists and leveraging the latest technology, NBR bridges the academic, business, and policy arenas. The institution disseminates its research through briefings, publications, conferences, Congressional testimony, and email forums, and by collaborating with leading institutions worldwide. NBR also provides exceptional internship opportunities to graduate and undergraduate students for the purpose of attracting and training the next generation of Asia specialists. NBR was started in 1989 with a major grant from the Henry M. Jackson Foundation. Funding for NBR’s research and publications comes from foundations, corporations, individuals, the U.S. government, and from NBR itself. NBR does not conduct proprietary or classified research. The organization undertakes contract work for government and private-sector organizations only when NBR can maintain the right to publish findings from such work. To download issues of the NBR publications, please visit the NBR website http://www.nbr.org. -

Joseph Harold Greenberg
JOSEPH HAROLD GREENBERG CORRECTED VERSION* Joseph H. Greenberg, one of the most original and influential linguists of the twentieth century, died at his home in Stanford, California, on May 7th, 2001, three weeks before his eighty-sixth birthday. Greenberg was a major pioneer in the development of linguistics as an empirical science. His work was always founded directly on quantitative data from a single language or from a wide range of languages. His chief legacy to contemporary linguistics is in the development of an approach to the study of language—typology and univerals—and to historical linguistics. Yet he also made major contributions to sociolinguistics, psycholinguistics, phonetics and phonology, morphology, and especially African language studies. Joe Greenberg was born on May 28th, 1915, in Brooklyn, New York, the second of two children. His father was a Polish Jew and his mother, a German Jew. His father’s family name was originally Zyto, but in one of those turn-of-the- century immigrant stories, he ended up taking the name of his landlord. Joe Greenberg’s early loves were music and languages. As a child he sat fascinated next to his mother while she played the piano, and asked her to teach him. She taught him musical notation and then found him a local teacher. Greenberg ended up studying with a Madame Vangerova, associated with the Curtis Institute of Music. Greenberg even gave a concert at Steinway Hall at the age of 14, and won a city-wide prize for best chamber music ensemble. But after finishing high school, Greenberg chose an academic career instead of a musical one, although he continued to play the piano every evening until near the end of his life. -

"Evolution of Human Languages": Current State of Affairs
«Evolution of Human Languages»: current state of affairs (03.2014) Contents: I. Currently active members of the project . 2 II. Linguistic experts associated with the project . 4 III. General description of EHL's goals and major lines of research . 6 IV. Up-to-date results / achievements of EHL research . 9 V. A concise list of actual problems and tasks for future resolution. 18 VI. EHL resources and links . 20 2 I. Currently active members of the project. Primary affiliation: Senior researcher, Center for Comparative Studies, Russian State University for the Humanities (Moscow). Web info: http://ivka.rsuh.ru/article.html?id=80197 George Publications: http://rggu.academia.edu/GeorgeStarostin Starostin Research interests: Methodology of historical linguistics; long- vs. short-range linguistic comparison; history and classification of African languages; history of the Chinese language; comparative and historical linguistics of various language families (Indo-European, Altaic, Yeniseian, Dravidian, etc.). Primary affiliation: Visiting researcher, Santa Fe Institute. Formerly, professor of linguistics at the University of Melbourne. Ilia Publications: http://orlabs.oclc.org/identities/lccn-n97-4759 Research interests: Genetic and areal language relationships in Southeast Asia; Peiros history and classification of Sino-Tibetan, Austronesian, Austroasiatic languages; macro- and micro-families of the Americas; methodology of historical linguistics. Primary affiliation: Senior researcher, Institute of Slavic Studies, Russian Academy of Sciences (Moscow / Novosibirsk). Web info / publications list (in Russian): Sergei http://www.inslav.ru/index.php?option- Nikolayev =com_content&view=article&id=358:2010-06-09-18-14-01 Research interests: Comparative Indo-European and Slavic studies; internal and external genetic relations of North Caucasian languages; internal and external genetic relations of North American languages (Na-Dene; Algic; Mosan). -

North Caucasus: Views from Within People’S Perspectives on Peace and Security
REPORT North Caucasus: views from within People’s perspectives on peace and security March 2012 North Caucasus: views from within People’s perspectives on peace and security SAFERWORLD MARCH 2012 Acknowledgements This report was written by Anna Matveeva, Honorary University Fellow, University of Exeter, with contributions from Igor Savin, Research Fellow, the Institute of Oriental Studies, Russian Academy of Sciences. It was edited by Craig Oliphant (Senior Advisor, Saferworld). Editorial contributions and comments were also provided by Luitgard Hammerer (Head of Europe and Central Asia, Saferworld). The report draws on participatory research conducted in five North Caucasus republics by Zhanna Khamdokhova (Kabardino-Balkaria), Igor Dulaev (North Ossetia), Musa Yusupov (Chechnya), Akhmet Yarlykapov (Dagestan), Anna Matveeva and Igor Savin (Ingushetia). The methodology for the research was developed by Anna Matveeva and Igor Savin with contributions from the Institute of Oriental Studies, Saferworld and the four researchers from the North Caucasus. For further reading, case studies from the individual republics can be accessed on the Saferworld website: www.saferworld.org.uk/PPP/chechnya www.saferworld.org.uk/PPP/northossetia www.saferworld.org.uk/PPP/kabardino-balkaria www.saferworld.org.uk/PPP/ingushetia www.saferworld.org.uk/PPP/dagestan The republic chapters were translated from Russian into English by Sophia Matveeva. John Newman copy-edited the report. The research was a collaboration between Saferworld and the Institute of Oriental Studies of the Russian Academy of Sciences. The People’s Peacemaking Perspectives project The People’s Peacemaking Perspectives project is a joint initiative implemented by Conciliation Resources and Saferworld and financed under the European Commission’s Instrument for Stability. -

Republic of Tatarstan 15 I
1 CONTENTS ABOUT AUTHORS 3 EXECUTIVE SUMMARY 4 INTRODUCTION 10 THE REPUBLIC OF TATARSTAN 15 I. POLITICAL ELITE 15 1. Vertical power structure 19 2. Governance model during the period of the President M. Shaimiev 20 3. Governance model during the period of the President R. Minnikhanov 22 4. Security forces as part of a consolidated project 27 5. Export of elites 28 II. PRESERVATION OF ETHNO-CULTURAL IDENTITY 30 1.The Tatar national movement 30 2. The Russian national movement 34 3. Language policy in Tatarstan 37 4. Results of post-Soviet language policy 47 5. Conclusion 50 THE REPUBLIC OF DAGESTAN 51 I. DAGESTAN ELITES AND THE FEDERAL GOVERNMENT 51 1. Birth of «clans» 53 2. Adaptation to the growing influence of Moscow 56 3. Mukhu Aliev: attempt to be equidistant from clans 58 4. Elite and the Caucasus Emirate 62 5. Return of the «levashintsy» and attempt at a civil dialogue 64 6. First attempt to eliminate clans 66 II. «EXTERNAL GOVERNANCE» 70 III. PRESERVATION OF ETHNO-CULTURAL IDENTITY 79 1. National movements and conflicts 79 2. Preservation of national languages 82 3. Conclusion 91 FINAL CONCLUSIONS 93 2 ABOUT AUTHORS Dr. Ekaterina SOKIRIANSKAIA is the founder and director at Conflict analysis and prevention center. From 2011 to 2017, she served as International Crisis Group’s Russia/North Caucasus Project Director, supervising the organisation’s research and advocacy in the region. From 2008-2011, Sokirianskaia established and supervised the work of Human rights Center Memorial’s regional offices in Kabardino-Balkariya and Dagestan. Before that, from 2003-2008 Sokirianskaia was permanently based in Ingushetia and Chechnya and worked as a researcher and projects director for Memorial and as an assistant professor at Grozny State University. -
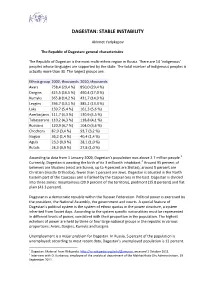
Dagestan: Stable Instability
DAGESTAN: STABLE INSTABILITY Akhmet Yarlykapov The Republic of Dagestan: general characteristics The Republic of Dagestan is the most multi-ethnic region in Russia. There are 14 ‘indigenous’ peoples whose languages are supported by the state. The total number of indigenous peoples is actually more than 30. The largest groups are: Ethnic group 2002, thousands 2010, thousands Avars 758,4 (29,4 %) 850,0 (29,4 %) Dargins 425,5 (16,5 %) 490,4 (17,0 %) Kumyks 365,8 (14,2 %) 431,7 (14,9 %) Lezgins 336,7 (13,1 %) 385,2 (13,3 %) Laks 139,7 (5,4 %) 161,3 (5,6 %) Azerbaijanis 111,7 (4,3 %) 130,9 (4,5 %) Tabassarans 110,2 (4,3 %) 118,8 (4,1 %) Russians 120,9 (4,7 %) 104,0 (3,6 %) Chechens 87,9 (3,4 %) 93,7 (3,2 %) Nogais 36,2 (1,4 %) 40,4 (1,4 %) Aguls 23,3 (0,9 %) 28,1 (1,0 %) Rutuls 24,3 (0,9 %) 27,8 (1,0 %) According to data from 1 January 2009, Dagestan’s population was above 2.7 million people. 1 Currently, Dagestan is awaiting the birth of its 3 millionth inhabitant. 2 Around 95 percent of believers are Muslims (most are Sunnis, up to 4 percent are Shiites), around 5 percent are Christian (mostly Orthodox), fewer than 1 percent are Jews. Dagestan is situated in the North Eastern part of the Caucasus and is flanked by the Caspian Sea in the East. Dagestan is divided into three zones: mountainous (39.9 percent of the territory), piedmont (15.8 percent) and flat plain (43.3 percent). -

Russia's Dagestan: Conflict Causes
RUSSIA’S DAGESTAN: CONFLICT CAUSES Europe Report N°192 – 3 June 2008 TABLE OF CONTENTS EXECUTIVE SUMMARY ...................................................................................................... i I. INTRODUCTION ............................................................................................................. 1 II. A FRAGILE INTER-ETHNIC BALANCE.................................................................... 2 A. INTER-ETHNIC COMPETITION OVER LAND AND STATE POSITIONS...............................................2 B. THE 2007 ELECTIONS .................................................................................................................4 1. Removing inter-ethnic competition from electoral politics..................................................4 2. Electoral violence and results ...............................................................................................5 III. ISLAMISM IN DAGESTAN AND CHECHEN CONNECTIONS.............................. 6 A. CHECHEN AND DAGESTANI ISLAMISTS IN THE 1990S .................................................................6 B. THE “HUNT FOR THE WAHHABIS” SINCE 1999 ...........................................................................8 C. SHARIAT JAMAAT’S GROWING INFLUENCE .................................................................................8 D. RENEWED TENSIONS WITH CHECHNYA .....................................................................................10 IV. VIOLENCE AGAINST STATE AUTHORITIES ...................................................... -

Long-Distance Reflexivization and Logophoricity in the Dargin Language Muminat Kerimova Florida International University
Florida International University FIU Digital Commons MA in Linguistics Final Projects College of Arts, Sciences & Education 2017 Long-Distance Reflexivization and Logophoricity in the Dargin Language Muminat Kerimova Florida International University Follow this and additional works at: https://digitalcommons.fiu.edu/linguistics_ma Part of the Linguistics Commons Recommended Citation Kerimova, Muminat, "Long-Distance Reflexivization and Logophoricity in the Dargin Language" (2017). MA in Linguistics Final Projects. 3. https://digitalcommons.fiu.edu/linguistics_ma/3 This work is brought to you for free and open access by the College of Arts, Sciences & Education at FIU Digital Commons. It has been accepted for inclusion in MA in Linguistics Final Projects by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. FLORIDA INTERNATIONAL UNIVERSITY Miami, Florida LONG-DISTANCE REFLEXIVIZATION AND LOGOPHORICITY IN THE DARGIN LANGUAGE A thesis submitted in partial fulfillment of the requirements for the degree of MASTER OF ARTS in LINGUISTICS by Muminat Kerimova 2017 ABSTRACT OF THE THESIS LONG-DISTANCE REFLEXIVIZATION AND LOGOPHORICITY IN THE DARGIN LANGUAGE by Muminat Kerimova Florida International University, 2017 Miami, Florida Professor Ellen Thompson, Major Professor The study of anaphora challenges us to determine the conditions under which the pronouns of a language are associated with possible antecedents. One of the theoretical questions is whether the distribution of pronominal forms is best explained by a syntactic, semantic or discourse level analysis. A more practical question is how we distinguish between anaphoric elements, e.g. what are the borders between the notions of pronouns, locally bound reflexives and long-distance reflexives? The study analyzes the anaphora device saj in Dargin that is traditionally considered to be a long-distance reflexivization language.