Genetic Determination of the Desert Massasauga Distribution in Texas
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
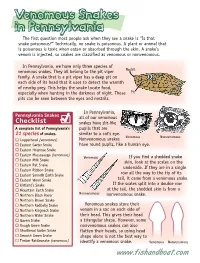
Venomous Snakes in Pennsylvania the First Question Most People Ask When They See a Snake Is “Is That Snake Poisonous?” Technically, No Snake Is Poisonous
Venomous Snakes in Pennsylvania The first question most people ask when they see a snake is “Is that snake poisonous?” Technically, no snake is poisonous. A plant or animal that is poisonous is toxic when eaten or absorbed through the skin. A snake’s venom is injected, so snakes are classified as venomous or nonvenomous. In Pennsylvania, we have only three species of venomous snakes. They all belong to the pit viper Nostril family. A snake that is a pit viper has a deep pit on each side of its head that it uses to detect the warmth of nearby prey. This helps the snake locate food, especially when hunting in the darkness of night. These Pit pits can be seen between the eyes and nostrils. In Pennsylvania, Pennsylvania Snakes all of our venomous Checklist snakes have slit-like A complete list of Pennsylvania’s pupils that are 21 species of snakes. similar to a cat’s eye. Venomous Nonvenomous Copperhead (venomous) Nonvenomous snakes Eastern Garter Snake have round pupils, like a human eye. Eastern Hognose Snake Eastern Massasauga (venomous) Venomous If you find a shedded snake Eastern Milk Snake skin, look at the scales on the Eastern Rat Snake underside. If they are in a single Eastern Ribbon Snake Eastern Smooth Earth Snake row all the way to the tip of its Eastern Worm Snake tail, it came from a venomous snake. Kirtland’s Snake If the scales split into a double row Mountain Earth Snake at the tail, the shedded skin is from a Northern Black Racer Nonvenomous nonvenomous snake. -

Ecology of the Desert Massasauga Rattlesnake in Colorado: Habitat and Resource Utilization
Ecology of the Desert Massasauga Rattlesnake in Colorado: Habitat and Resource Utilization A report to the CDOW and USFWS for the Colorado Wildlife Conservation Grant Program Dr. Stephen P. Mackessy School of Biological Sciences University of Northern Colorado 501 20th St., CB 92 Greeley, CO 80639-0017 USA Tel: (970)-351-2429 Fax: (970)-351-2335 Email: [email protected] http://www.unco.edu/nhs/biology/faculty_staff/mackessy_stephen.htm Desert Massasauga (Sistrurus catenatus edwardsii) from Lincoln County, Colorado ABSTRACT We conducted a radiotelemetric study on a population of Desert Massasaugas (Sistrurus catenatus edwardsii) in Lincoln County, SE Colorado. Massasaugas were most active between 14 and 30 °C, with an average ambient temperature during activity of 22.1°C. In the spring, snakes make long distance movements (up to 2 km) from the hibernaculum (shortgrass, compacted clay soils) to summer foraging areas (mixed grass/sandsage, sand hills). Summer activity is characterized by short distance movements, and snakes are most often observed at the base of sandsage in ambush or resting coils. Massasaugas gave birth to 5-7 young in late August, and reproduction appears to be biennial. Observations on three radioed gravid females indicated that Desert Massasaugas show maternal attendance for at least five days post-parturition. Snakes returned to the hibernaculum area in October and appear to hibernate individually in rodent burrows most commonly. However, the immediate region is utilized by several other species of snakes, particularly Prairie Rattlesnakes (Crotalus viridis viridis), and Massasaugas were also observed at the entrance to these den sites, suggesting that they are used by the Massasaugas as well. -

The Eastern Massasauga Rattlesnake Is Toxic, but Only a Small Amount Is Injected Through Short Fangs
What’s so special about the massasauga? Northern Indiana’s The rare eastern massasauga is northern Indiana’s only native rattlesnake. Endangered Because eastern massasaugas are shy and secretive, you may never see one in the wild. These snakes hide under Rattlesnake brush and retreat to a sheltered area if spotted in the open. Although eastern massasaugas are venomous, they usually flee from humans rather than bite. Their venom Range of the Eastern Massasauga Rattlesnake is toxic, but only a small amount is injected through short fangs. The last human fatality from an eastern Why are they endangered? massasauga bite occurred more than 60 years ago. Eastern massasaugas are endangered over much of their Eastern massasaugas live in range because their wetland habitats are often drained and northern Indiana’s swamps, Eastern massasaugas play an important role in the filled for development. These snakes are also collected for bogs, and wetlands. ecosystem by feeding on mice, voles, and shrews, thus the illegal reptile trade. People may kill massasaugas out of Christopher Smith helping to keep the rodent population under control. fear, not realizing the snakes’ importance to the ecosystem. People and snakes can live together. Nicholas Scobel Understanding snakes is the first step. Learn to identify eastern massasaugas and other Indiana snakes. A field guide to native reptiles can help. Wear proper footwear when hiking in areas where snakes might be found, especially at night. Stay on trails. Limit the use of pesticides and other chemicals on natural areas on your property. All wildlife will benefit. Teach your family and friends about snakes and what to do if they see an eastern massasauga. -

Jennifer Szymanski Usfish and Wildlife Service Endangered
Written by: Jennifer Szymanski U.S.Fish and Wildlife Service Endangered Species Division 1 Federal Drive Fort Snelling, Minnesota 55111 Acknowledgements: Numerous State and Federal agency personnel and interested individuals provided information regarding Sistrurus c. catenatus’status. The following individuals graciously provided critical input and numerous reviews on portions of the manuscript: Richard Seigel, Robert Hay, Richard King, Bruce Kingsbury, Glen Johnson, John Legge, Michael Oldham, Kent Prior, Mary Rabe, Andy Shiels, Doug Wynn, and Jeff Davis. Mary Mitchell and Kim Mitchell provided graphic assistance. Cover photo provided by Bruce Kingsbury Table of Contents Taxonomy....................................................................................................................... 1 Physical Description....................................................................................................... 3 Distribution & State Status............................................................................................. 3 Illinois................................................................................................................. 5 Indiana................................................................................................................ 5 Iowa.................................................................................................................... 5 Michigan............................................................................................................ 6 Minnesota.......................................................................................................... -

The Massasauga Rattlesnake
Pennsylvania Amphibians & Reptiles TTheh e MMassasauga s s a s a u g a R a t t l e s n a k e by Andrew L. Shiels “Massasauga,” “swamp rattler,” and “black snapper,” are a 24 to 30 inches. It has a grayish body color with a series of few of the nicknames given to a rare, secretive, yet interest- dark-brown, irregularly shaped blotches along the top of ing rattlesnake found in northwestern Pennsylvania. Its the back. These blotches are the source of the Latin spe- limited distribution and unique ecological needs have un- cies name “catenatus,” which means “chain-like.” Along fortunately placed this species among Pennsylvania’s most the sides of the body are two to three rows of smaller, more endangered. Out of sight and out of mind to most Penn- rounded spots. sylvanians, the massasauga has been quietly disappearing Massasaugas are pit vipers, which means there is a for years. However, recent conservation efforts have been heat-sensing pit on the side of the head between the eye undertaken to ensure its survival in the Commonwealth. and nostril. Like our other two venomous species, the Understanding its biology and ecology are keys to devel- northern copperhead and the eastern timber rattlesnake, oping an effective conservation strategy. massasaugas have vertically elliptical pupils and a single The eastern massasauga (Sistrurus catenatus catenatus) row of scales on the underside of the tail. Unlike timber is a medium-sized venomous snake that reaches lengths of rattlesnakes, which have many small scales on the head, a massasauga’s head is covered with nine large, scale plates typical of our non-venomous species. -

Lesser Known Snakes (PDF)
To help locate food, snakes repeat- edly flick out their narrow forked tongues to bring odors to a special sense organ in their mouths. Snakes are carnivores, eating a variety of items, including worms, with long flexible bodies, snakes insects, frogs, salamanders, fish, are unique in appearance and small birds and mammals, and even easily recognized by everyone. other snakes. Prey is swallowed Unfortunately, many people are whole. Many people value snakes afraid of snakes. But snakes are for their ability to kill rodent and fascinating and beautiful creatures. insect pests. Although a few snakes Their ability to move quickly and are venomous, most snakes are quietly across land and water by completely harmless to humans. undulating their scale-covered Most of the snakes described bodies is impressive to watch. here are seldom encountered— Rather than moveable eyelids, some because they are rare, but snakes have clear scales covering most because they are very secre- their eyes, which means their eyes tive, usually hiding under rocks, are always open. Some snake logs or vegetation. For information species have smooth scales, while on the rest of New York’s 17 species others have a ridge on each scale of snakes, see the August 2001 that gives it a file-like texture. issue of Conservationist. Text by Alvin Breisch and Richard C. Bothner, Maps by John W. Ozard (based on NY Amphibian & Reptile Atlas Project) Artwork by Jean Gawalt, Design by Frank Herec This project was funded in part by Return A Gift To Wildlife tax check-off, U.S. Fish & Wildlife Service and the NYS Biodiversity Research Institute. -

Eastern Massasauga Natural Diversity Section June 2011
Species Action Plan Eastern Massasauga Natural Diversity Section June 2011 Species Action Plan: Eastern Massasauga (Sistrurus catenatus catenatus) Purpose: This plan provides an initial five year blueprint for the actions needed to attain near-term and, ultimately, long-term goals for the conservation and recovery of the eastern massasauga. The action plan is Figure 1. Eastern Massasauga (Sistrurus a living document and will be updated as catenatus). Photo-Charlie Eichelberger. needed to reflect progress toward those goals and to incorporate new information as it becomes available. Description: A small, stout-bodied rattlesnake. The body is patterned with a Goals: The immediate goal is to maintain series of large, dark brown, mid-dorsal the extant populations of eastern blotches and two to three rows of lateral massasauaga in the Commonwealth and to blotches set upon a light gray or tan protect its remaining habitat. The ground color. The tail has three to six dark secondary goal is to enhance extant cross-bands. The venter is typically black. populations by improving and increasing Neonates have the same pattern and local habitat. The long-term recovery goal coloration as adults, with the exception of is to increase viable, reproducing the neonate exhibiting a yellow tip at the populations in extant and historic locations proximate end of the tail. At birth, S. c. with the possibility of removal of the catenatus average 23.6 cm in total length species from the Pennsylvania list of (Reinert, 1981) and have a single rattle endangered species (58 Pa. Code §75.1). segment (button) at the tip of their tail. -

Western Massasauga ( ЛХЦФЧФЧХ ЦЗФЙЗПЛРЧХ)
Western Massasauga (Sistrurus tergeminus ) A Species Conservation Assessment for The Nebraska Natural Legacy Project Prepared by Melissa J. Panella and Brent D. Johnson for Nebraska Game and Parks Commission Wildlife Division Lincoln, Nebraska June 2014 The mission of the Nebraska Natural Legacy Project is to implement a blueprint for conserving Nebraska’s flora, fauna and natural habitats through the proactive, voluntary conservation actions of partners, communities and individuals. Purpose The primary goal in development of at-risk species conservation assessments is to compile biological and ecological information that may assist conservation practitioners in making decisions regarding the conservation of species of interest. The Nebraska Natural Legacy Project recognizes the Western Massasauga ( Sistrurus tergeminus ) as a Tier I at-risk species. Provided are some general management recommendations regarding Western Massasaugas. Conservation practitioners will need to use professional judgment to make specific management decisions based on objectives, location, and a multitude of variables. This resource was designed to share available knowledge of this at-risk species that will aid in the decision-making process or in identifying research needs to benefit the species. Species conservation assessments will need to be updated as relevant scientific information becomes available and/or conditions change. Though the Nebraska Natural Legacy Project focuses efforts in the state’s Biologically Unique Landscapes, it is recommended that whenever possible, practitioners make considerations for a species throughout its range in order to increase the outcome of successful conservation efforts. And in the case of conservation for massasaugas, it is particularly necessary to take into account the seasonal needs of the species and conserve both wintering and summer foraging habitat. -

Desert Massasauga Rattlesnake (Sistrurus Catenatus Edwardsii): a Technical Conservation Assessment
Desert Massasauga Rattlesnake (Sistrurus catenatus edwardsii): A Technical Conservation Assessment Prepared for the USDA Forest Service, Rocky Mountain Region, Species Conservation Project December 12, 2005 Stephen P. Mackessy, Ph.D.1 with input from Douglas A. Keinath2, Mathew McGee2, Dr. David McDonald3, and Takeshi Ise3 1 School of Biological Sciences, University of Northern Colorado, 501 20th St., CB 92, Greeley, CO 80639-0017 2 Wyoming Natural Diversity Database, University of Wyoming, P.O. Box 3381, Laramie, WY 82071 3 Department of Zoology and Physiology, University of Wyoming, P.O. Box 3166, Laramie, WY 82071 Peer Review Administered by Society for Conservation Biology Mackessy, S.P. (2005, December 12). Desert Massasauga Rattlesnake (Sistrurus catenatus edwardsii): a technical conservation assessment. [Online]. USDA Forest Service, Rocky Mountain Region. Available: http:// www.fs.fed.us/r2/projects/scp/assessments/massasauga.pdf [date of access]. ACKNOWLEDGMENTS We would like to thank the members of the massasauga listserv for information provided, including but not limited to Tom Anton, Gary Casper, Frank Durbian, Andrew Holycross, Rebecca Key, David Mauger, Jennifer Szymanski, and Darlene Upton. For excellent field work on surveys of southeastern Colorado and radiotelemetry of desert massasaugas, the lead author thanks Enoch Bergman, Ron Donoho, Ben Hill, Justin Hobert, Rocky Manzer, Chad Montgomery, James Siefert, Kevin Waldron, and Andrew Wastell. John Palmer and the Palmer family generously provided access to their ranch, which was critical for both survey and telemetry work, and many other kindnesses. For discussions about massasaugas at various times, I thank David Chiszar, Harry Greene, Geoff Hammerson, Andrew Holycross, Lauren Livo, Richard Seigel, and Hobart M. -

EASTERN MASSASAUGA Sistrurus Catenatus Catenatus Identification, Status, Ecology, and Conservation in Midwest
EASTERN MASSASAUGA Sistrurus catenatus catenatus Identification, Status, Ecology, and Conservation in Midwest IDENTIFICATION General Appearance Characteristic of all rattlesnake species, the Eastern Massasauga has a rattle on the end of its tail and heat sensing pits situated between the eyes and nostrils (see photo next page). The background color of the Eastern Massasauga is typically gray or brownish gray, with a row of dark-brown to black dorsal blotches and Eastern Massasaugas typically have a light brown or gray three rows of small brown to background color with darker brown or gray blotches black spots on each side of the intermixed. Photo by A. Fortin. body. The dorsal or side blotches in some individuals may be connected, giving the appearance of stripes. Some Eastern Massasaugas may be black, or nearly so, in some areas. In this melanistic phase a pattern is not readily discernable. Apart from the tail and rattle, juvenile Eastern Massasaugas appear very similar to adult snakes. The tails of juveniles are bright yellow, and lack a fully formed rattle: instead, they are born with a “button” on the tips of their tails. Eastern Massasaugas are considered medium-sized rattlesnakes with adults reaching Massasaugas may be melanistic (black). lengths of up to one meter. Photo by G. Lipps, Jr. Subspecies The Eastern Massasauga is one of three subspecies of S. catenatus. The other two subspecies are the Western Massasauga (S. c. tergeminus) and the Desert Massasauga (S. c. edwardsii). Some controversy surrounds the correct taxonomic arrangement of these snakes. Perhaps ultimately they will be viewed as two or three species. -

Thirty-Nine Species of Snakes Inhabit Illinois, Dwelling in Forests, Grasslands
I l l i n o i s yellowbelly water snake Nerodia erythrogaster SNAKESSNAKES eastern hognose snake Heterodon platirhinos eastern worm snake Carphophis amoenus racer Coluber constrictor western ribbon snake Thamnophis proximus Graham’s crayfish snake Regina grahamii northern water snake Nerodia sipedon western mud snake Farancia abacura smooth earth snake Virginia valeriae common garter snake Thamnophis sirtalis redbelly snake Storeria occipitomaculata western fox snake Pantherophis vulpinus prairie kingsnake Lampropeltis calligaster bullsnake Pituophis catenifer diamondback water snake Nerodia rhombifer common kingsnake Lampropeltis getula red milk snake Lampropeltis triangulum syspila brown snake Storeria dekayi rough green snake Opheodrys aestivus black rat snake juvenile cottonmouth juvenile ringneck snake Diadophis punctatus black rat snake Pantherophis spiloides cottonmouth Agkistrodon piscivorus timber rattlesnake juvenile copperhead juvenile eastern massasauga juvenile timber rattlesnake Crotalus horridus copperhead Agkistrodon contortrix eastern massasauga Sistrurus catenatus Species List Family Colubridae This poster was made possible by: eastern worm snake Carphophis amoenus hirty-nine species of snakes inhabit Illinois, dwelling in forests, grasslands, marshes, swamps, ponds, racer Coluber constrictor ringneck snake Diadophis punctatus western mud snake Farancia abacura Illinois Department of Natural Resources lakes, streams, rivers, and sloughs. Some species are quite common, while others are very rare. These eastern hognose snake Heterodon platirhinos prairie kingsnake Lampropeltis calligaster Division of Education common kingsnake Lampropeltis getula Division of Natural Heritage reptiles are solitary predators that eat a variety of prey. Snakes have interesting structural features Classification: red milk snake Lampropeltis triangulum syspila T yellowbelly water snake Nerodia erythrogaster Illinois State Museum Kingdom Animalia diamondback water snake Nerodia rhombifer including the Jacobson’s organ, which is used to detect odors. -

Venomous Snakes in Pennsylvania
Coexisting with snakes Snakes are an essential component of Pennsylvania’s wildlife What are the Differences? resources. Fear of snakes often stems from a lack of knowledge of its habits and role in the ecosystem. Some people don’t think about Pennsylvania snakes until snakes show up in their backyard, shed or house. The Venomous or nonvenomous? majority of snakes appearing in these areas are nonvenomous and There are 18 nonvenomous and three venomous snake harmless. Still, it is understandable that when a snake is discovered Snakes species in Pennsylvania. Some key physical differences between near or in the house, people quickly seek a way to remove it. these two groups of Pennsylvania snakes facilitate identification. Take precautions to prevent snakes from entering dwellings. All Pennsylvania native venomous snakes possess an Sealing openings and cracks, especially at or near ground level, is indentation or pit on each side of the head between the eye and an excellent preventive measure. Removing debris in the yard or nostril, a vertically elliptical eye pupil resembling that of a cat, and moving the wood pile farther from the house can also help. Snakes a single row of scales on the underside of the tail. have biological needs such as food and cover that must be met Nonvenomous Pennsylvania native snakes do not possess for them to remain in a location. If those needs are not met, they facial pits, have round eye pupils and a double row of scales on move on. the underside of the tail. Field guides are great tools for bridging gaps in knowledge.