Millipede Genomes Reveal Unique Adaptation of Genes And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Integrative Revision of the Giant Pill-Millipede Genus Sphaeromimus
A peer-reviewed open-access journal ZooKeys 414: 67–107 (2014) New Sphaeromimus species from Madagascar 67 doi: 10.3897/zookeys.414.7730 RESEARCH ARTICLE www.zookeys.org Launched to accelerate biodiversity research Integrative revision of the giant pill-millipede genus Sphaeromimus from Madagascar, with the description of seven new species (Diplopoda, Sphaerotheriida, Arthrosphaeridae) Thomas Wesener1,2,†, Daniel Minh-Tu Le1,3,‡, Stephanie F. Loria1,4,§ 1 Field Museum of Natural History, Zoology - Insects, 1400 S. Lake Shore Drive, 60605 Chicago, Illinois, U.S.A. 2 Zoologisches Forschungsmuseum Alexander Koenig, Leibniz Institute for Animal Biodiversity, Center for Taxonomy and Evolutionary Research (Section Myriapoda), Adenauerallee 160, 53113 Bonn, Germany 3 School of the Art Institute of Chicago, 36 S. Wabash Avenue, 60603 Chicago, Illinois, U.S.A. 4 American Museum of Natural History, Richard Glider Graduate School, Central Park West at 79th Street, New York, U.S.A. † http://zoobank.org/86DEA7CD-988C-43EC-B9D6-C51000595B47 ‡ http://zoobank.org/AD76167C-3755-4803-AEB5-4CD9A7CB820A § http://zoobank.org/ED92B15A-10F9-47B8-A8FA-D7673007F8A5 Corresponding author: Thomas Wesener ([email protected]) Academic editor: D.V. Spiegel | Received 15 April 2014 | Accepted 8 May 2014 | Published 6 June 2014 http://zoobank.org/59FA2886-34C2-4AEF-9783-3347E5EBC702 Citation: Wesener T, Le DM-T, Loria SF (2014) Integrative revision of the giant pill-millipede genus Sphaeromimus from Madagascar, with the description of seven new species (Diplopoda, Sphaerotheriida, Arthrosphaeridae). ZooKeys 414: 67–107. doi: 10.3897/zookeys.414.7730 Abstract The Malagasy giant pill-millipede genusSphaeromimus de Saussure & Zehntner, 1902 is revised. Seven new species, S. -

Supra-Familial Taxon Names of the Diplopoda Table 4A
Milli-PEET, Taxonomy Table 4 Page - 1 - Table 4: Supra-familial taxon names of the Diplopoda Table 4a: List of current supra-familial taxon names in alphabetical order, with their old invalid counterpart and included orders. [Brackets] indicate that the taxon group circumscribed by the old taxon group name is not recognized in Shelley's 2003 classification. Current Name Old Taxon Name Order Brannerioidea in part Trachyzona Verhoeff, 1913 Chordeumatida Callipodida Lysiopetalida Chamberlin, 1943 Callipodida [Cambaloidea+Spirobolida+ Chorizognatha Verhoeff, 1910 Cambaloidea+Spirobolida+ Spirostreptida] Spirostreptida Chelodesmidea Leptodesmidi Brölemann, 1916 Polydesmida Chelodesmidea Sphaeriodesmidea Jeekel, 1971 Polydesmida Chordeumatida Ascospermophora Verhoeff, 1900 Chordeumatida Chordeumatida Craspedosomatida Jeekel, 1971 Chordeumatida Chordeumatidea Craspedsomatoidea Cook, 1895 Chordeumatida Chordeumatoidea Megasacophora Verhoeff, 1929 Chordeumatida Craspedosomatoidea Cheiritophora Verhoeff, 1929 Chordeumatida Diplomaragnoidea Ancestreumatoidea Golovatch, 1977 Chordeumatida Glomerida Plesiocerata Verhoeff, 1910 Glomerida Hasseoidea Orobainosomidi Brolemann, 1935 Chordeumatida Hasseoidea Protopoda Verhoeff, 1929 Chordeumatida Helminthomorpha Proterandria Verhoeff, 1894 all helminthomorph orders Heterochordeumatoidea Oedomopoda Verhoeff, 1929 Chordeumatida Julida Symphyognatha Verhoeff, 1910 Julida Julida Zygocheta Cook, 1895 Julida [Julida+Spirostreptida] Diplocheta Cook, 1895 Julida+Spirostreptida [Julida in part[ Arthrophora Verhoeff, -

Millipedes and Centipedes
MILLIPEDES AND CENTIPEDES Integrated Pest Management In and Around the Home sprouting seeds, seedlings, or straw- berries and other ripening fruits in contact with the ground. Sometimes individual millipedes wander from their moist living places into homes, but they usually die quickly because of the dry conditions and lack of food. Occasionally, large (size varies) (size varies) numbers of millipedes migrate, often uphill, as their food supply dwindles or their living places become either Figure 1. Millipede (left); Centipede (right). too wet or too dry. They may fall into swimming pools and drown. Millipedes and centipedes (Fig. 1) are pedes curl up. The three species When disturbed they do not bite, but often seen in and around gardens and found in California are the common some species exude a defensive liquid may be found wandering into homes. millipede, the bulb millipede, and the that can irritate skin or burn the eyes. Unlike insects, which have three greenhouse millipede. clearly defined body sections and Life Cycle three pairs of legs, they have numer- Millipedes may be confused with Adult millipedes overwinter in the ous body segments and numerous wireworms because of their similar soil. Eggs are laid in clutches beneath legs. Like insects, they belong to the shapes. Wireworms, however, are the soil surface. The young grow largest group in the animal kingdom, click beetle larvae, have only three gradually in size, adding segments the arthropods, which have jointed pairs of legs, and stay underneath the and legs as they mature. They mature bodies and legs and no backbone. soil surface. -

Millipedes and Centipedes? Millipedes and Centipedes Are Both Arthropods in the Subphylum Myriapoda Meaning Many Legs
A Teacher’s Resource Guide to Millipedes & Centipedes Compiled by Eric Gordon What are millipedes and centipedes? Millipedes and centipedes are both arthropods in the subphylum Myriapoda meaning many legs. Although related to insects or “bugs”, they are not actually insects, which generally have six legs. How can you tell the difference between millipedes and centipedes? Millipedes have two legs per body segment and are typically have a body shaped like a cylinder or rod. Centipedes have one leg per body segment and their bodies are often flat. Do millipedes really have a thousand legs? No. Millipedes do not have a thousand legs nor do all centipedes have a hundred legs despite their names. Most millipedes have from 40-400 legs with the maximum number of legs reaching 750. No centipede has exactly 100 legs (50 pairs) since centipedes always have an odd number of pairs of legs. Most centipedes have from 30- 50 legs with one order of centipedes (Geophilomorpha) always having much more legs reaching up to 350 legs. Why do millipedes and centipedes have so many legs? Millipedes and centipedes are metameric animals, meaning that their body is divided into segments most of which are completely identical. Metamerization is an important phenomenon in evolution and even humans have a remnant of former metamerization in the repeating spinal discs of our backbone. Insects are thought to have evolved from metameric animals after specializing body segments for specific functions such as the head for sensation and the thorax for locomotion. Millipedes and centipedes may be evolutionary relatives to the ancestor of insects and crustaceans. -

A New Species of Illacme Cook & Loomis, 1928
A peer-reviewed open-access journal ZooKeys 626: 1–43A new (2016) species of Illacme Cook and Loomis, 1928 from Sequoia National Park... 1 doi: 10.3897/zookeys.626.9681 RESEARCH ARTICLE http://zookeys.pensoft.net Launched to accelerate biodiversity research A new species of Illacme Cook & Loomis, 1928 from Sequoia National Park, California, with a world catalog of the Siphonorhinidae (Diplopoda, Siphonophorida) Paul E. Marek1, Jean K. Krejca2, William A. Shear3 1 Virginia Polytechnic Institute and State University, Department of Entomology, Price Hall, Blacksburg, Virginia, USA 2 Zara Environmental LLC, 1707 W FM 1626, Manchaca, Texas, USA 3 Hampden-Sydney College, Department of Biology, Gilmer Hall, Hampden-Sydney, Virginia, USA Corresponding author: Paul E. Marek ([email protected]) Academic editor: R. Mesibov | Received 25 July 2016 | Accepted 19 September 2016 | Published 20 October 2016 http://zoobank.org/36E16503-BC2B-4D92-982E-FC2088094C93 Citation: Marek PE, Krejca JK, Shear WA (2016) A new species of Illacme Cook & Loomis, 1928 from Sequoia National Park, California, with a world catalog of the Siphonorhinidae (Diplopoda, Siphonophorida). ZooKeys 626: 1–43. doi: 10.3897/zookeys.626.9681 Abstract Members of the family Siphonorhinidae Cook, 1895 are thread-like eyeless millipedes that possess an astounding number of legs, including one individual with 750. Due to their cryptic lifestyle, rarity in natural history collections, and sporadic study over the last century, the family has an unclear phylogenetic placement, and intrafamilial relationships remain unknown. Here we report the discovery of a second spe- cies of Illacme, a millipede genus notable for possessing the greatest number of legs of any known animal on the planet. -

Some Aspects of the Ecology of Millipedes (Diplopoda) Thesis
Some Aspects of the Ecology of Millipedes (Diplopoda) Thesis Presented in Partial Fulfillment of the Requirements for the Degree Master of Science in the Graduate School of The Ohio State University By Monica A. Farfan, B.S. Graduate Program in Evolution, Ecology, and Organismal Biology The Ohio State University 2010 Thesis Committee: Hans Klompen, Advisor John W. Wenzel Andrew Michel Copyright by Monica A. Farfan 2010 Abstract The focus of this thesis is the ecology of invasive millipedes (Diplopoda) in the family Julidae. This particular group of millipedes are thought to be introduced into North America from Europe and are now widely found in many urban, anthropogenic habitats in the U.S. Why are these animals such effective colonizers and why do they seem to be mostly present in anthropogenic habitats? In a review of the literature addressing the role of millipedes in nutrient cycling, the interactions of millipedes and communities of fungi and bacteria are discussed. The presence of millipedes stimulates fungal growth while fungal hyphae and bacteria positively effect feeding intensity and nutrient assimilation efficiency in millipedes. Millipedes may also utilize enzymes from these organisms. In a continuation of the study of the ecology of the family Julidae, a comparative study was completed on mites associated with millipedes in the family Julidae in eastern North America and the United Kingdom. The goals of this study were: 1. To establish what mites are present on these millipedes in North America 2. To see if this fauna is the same as in Europe 3. To examine host association patterns looking specifically for host or habitat specificity. -

Accepted Manuscript Macroecological Inferences on Soil Fauna Through Comparative Niche Modeling: the Case of Hormogastridae (Annelida, Oligochaeta) Daniel F
This is an Open Access document downloaded from ORCA, Cardiff University's institutional repository: http://orca.cf.ac.uk/91465/ This is the author’s version of a work that was submitted to / accepted for publication. Citation for final published version: Marchán, Daniel F., Refoyo, Pablo, Fernández, Rosa, Novo Rodriguez, Marta, de Sosa, Irene and Díaz Cosín, Darío J. 2016. Macroecological inferences on soil fauna through comparative niche modeling: The case of Hormogastridae (Annelida, Oligochaeta). European Journal of Soil Biology 75 , pp. 115-122. 10.1016/j.ejsobi.2016.05.003 file Publishers page: http://dx.doi.org/10.1016/j.ejsobi.2016.05.003 <http://dx.doi.org/10.1016/j.ejsobi.2016.05.003> Please note: Changes made as a result of publishing processes such as copy-editing, formatting and page numbers may not be reflected in this version. For the definitive version of this publication, please refer to the published source. You are advised to consult the publisher’s version if you wish to cite this paper. This version is being made available in accordance with publisher policies. See http://orca.cf.ac.uk/policies.html for usage policies. Copyright and moral rights for publications made available in ORCA are retained by the copyright holders. Accepted manuscript Macroecological inferences on soil fauna through comparative niche modeling: the case of Hormogastridae (Annelida, Oligochaeta) Daniel F. Marchán1*#, Pablo Refoyo1#, Rosa Fernández2, Marta Novo3, Irene de Sosa1, Darío J. Díaz Cosín1 DOI: 10.1016/j.ejsobi.2016.05.003 To appear in: European Journal of Soil Biology Received date: 25 January 2016 Revised date: 12 May 2016 Accepted date: 17 May 2016 Please cite this article as: Marchán DF, Refoyo P, Fernández R, Novo M, de Sosa I, Díaz Cosín DJ (2016). -

The Millipedes and Centipedes of Chiapas Amber
14 4 ANNOTATED LIST OF SPECIES Check List 14 (4): 637–646 https://doi.org/10.15560/14.4.637 The millipedes and centipedes of Chiapas amber Francisco Riquelme1, Miguel Hernández-Patricio2 1 Laboratorio de Sistemática Molecular. Escuela de Estudios Superiores del Jicarero, Universidad Autónoma del Estado de Morelos, Jicarero C.P. 62909, Morelos, Mexico. 2 Subcoordinación de Inventarios Bióticos, Comisión Nacional para el Conocimiento y Uso de la Biodiversidad, Tlalpan C.P. 14010, Mexico City, Mexico. Corresponding author: Francisco Riquelme, [email protected] Abstract An inventory of fossil millipedes (class Diplopoda) and centipedes (class Chilopoda) from Miocene Chiapas amber, Mexico, is presented, with the inclusion of new records. For Diplopoda, 34 members are enumerated, for which 31 are described as new fossil records of the orders Siphonophorida Newport, 1844, Spirobolida Bollman, 1893, Polydesmida Leach, 1895, Stemmiulida Pocock, 1894, and the superorder Juliformia Attems, 1926. For Chilopoda 8 fossils are listed, for which 3 are new records of the order Geophilomorpha Pocock, 1895 and 2 are of the order Scolopendromorpha Pocock, 1895. Key words Miocene, Mexico, Diplopoda, Chilopoda. Academic editor: Peter Dekker | Received 14 May 2018 | Accepted 26 July 2018 | Published 10 August 2018 Citation: Riquelme F, Hernández-Patricio F (2018) The millipedes and centipedes of Chiapas amber. Check List 14 (4): 637–646. https://doi. org/10.15560/14.4.637 Introduction Diplopoda fossil record worldwide (Edgecombe 2015). Two other centipedes have been reported in Ross et al. The extant species of millipedes and centipedes distributed (2016). Other records of millipedes have been mentioned across Mexico have been studied since the initial reports in the literature but are questionable because of a lack of Brand (1839) and Persbosc (1839). -

Ordinal-Level Phylogenomics of the Arthropod Class
Ordinal-Level Phylogenomics of the Arthropod Class Diplopoda (Millipedes) Based on an Analysis of 221 Nuclear Protein-Coding Loci Generated Using Next- Generation Sequence Analyses Michael S. Brewer1,2*, Jason E. Bond3 1 Department of Environmental Science, Policy, and Management, University of California Berkeley, Berkeley, California, United States of America, 2 Department of Biology, East Carolina University, Greenville, North Carolina, United States of America, 3 Department of Biological Sciences and Auburn University Museum of Natural History, Auburn University, Auburn, Alabama, United States of America Abstract Background: The ancient and diverse, yet understudied arthropod class Diplopoda, the millipedes, has a muddled taxonomic history. Despite having a cosmopolitan distribution and a number of unique and interesting characteristics, the group has received relatively little attention; interest in millipede systematics is low compared to taxa of comparable diversity. The existing classification of the group comprises 16 orders. Past attempts to reconstruct millipede phylogenies have suffered from a paucity of characters and included too few taxa to confidently resolve relationships and make formal nomenclatural changes. Herein, we reconstruct an ordinal-level phylogeny for the class Diplopoda using the largest character set ever assembled for the group. Methods: Transcriptomic sequences were obtained from exemplar taxa representing much of the diversity of millipede orders using second-generation (i.e., next-generation or high-throughput) sequencing. These data were subject to rigorous orthology selection and phylogenetic dataset optimization and then used to reconstruct phylogenies employing Bayesian inference and maximum likelihood optimality criteria. Ancestral reconstructions of sperm transfer appendage development (gonopods), presence of lateral defense secretion pores (ozopores), and presence of spinnerets were considered. -
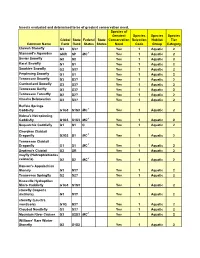
Insects Evaluated and Determined to Be of Greatest Conservation Need
Insects evaluated and determined to be of greatest conservation need. Species of Greatest Species Species Species Global State Federal State Conservation Selection Habitat Tier Common Name Rank Rank Status Status Need Code Group Category Etowah Stonefly G3 S3? Yes 1 Aquatic 2 Stannard's Agarodes GNR SP MC 1 Yes 1 Aquatic 2 Sevier Snowfly G2 S2 Yes 1 Aquatic 2 Karst Snowfly G1 S1 Yes 1 Aquatic 2 Smokies Snowfly G2 S3? Yes 1 Aquatic 2 Perplexing Snowfly G1 S1 Yes 1 Aquatic 2 Tennessee Snowfly G3 S3? Yes 1 Aquatic 2 Cumberland Snowfly G3 S3? Yes 1 Aquatic 2 Tennessee Sallfly G3 S3? Yes 1 Aquatic 2 Tennessee Forestfly G2 S2? Yes 1 Aquatic 2 Cheaha Beloneurian G3 S3? Yes 1 Aquatic 2 Buffalo Springs Caddisfly G1G3 S1S3 MC 1 Yes 1 Aquatic 2 Helma's Net-spinning Caddisfly G1G3 S1S3 MC 1 Yes 1 Aquatic 2 Sequatchie Caddisfly G1 S1 C Yes 1 Aquatic 2 Cherokee Clubtail Dragonfly G2G3 S1 MC 1 Yes 1 Aquatic 2 Tennessee Clubtail Dragonfly G1 S1 MC 1 Yes 1 Aquatic 2 Septima's Clubtail G2 SR Yes 1 Aquatic 2 mayfly (Habrophlebiodes celeteria) G2 S2 MC 1 Yes 1 Aquatic 2 Hanson's Appalachian Stonely G3 S3? Yes 1 Aquatic 2 Tennessee Springfly G2 S2? Yes 1 Aquatic 2 Knoxville Hydroptilan Micro Caddisfly G1G3 S1S3 Yes 1 Aquatic 2 stonefly (Isoperla distincta) G3 S3? Yes 1 Aquatic 2 stonefly (Leuctra monticola) G1Q S3? Yes 1 Aquatic 2 Clouded Needlefly G3 S3? Yes 1 Aquatic 2 Mountain River Cruiser G3 S2S3 MC 1 Yes 1 Aquatic 2 Williams' Rare Winter Stonefly G2 S1S2 Yes 1 Aquatic 2 Con't...Insects evaluated and determined to be of greatest conservation need. -

Zootaxa, Arthropoda: Diplopoda, Field Museum
ZOOTAXA 1005 The millipede type specimens in the Collections of the Field Museum of Natural History (Arthropoda: Diplopoda) PETRA SIERWALD, JASON E. BOND & GRZEGORZ T. GURDA Magnolia Press Auckland, New Zealand PETRA SIERWALD, JASON E. BOND & GRZEGORZ T. GURDA The millipede type specimens in the Collections of the Field Museum of Natural History (Arthro- poda: Diplopoda) (Zootaxa 1005) 64 pp.; 30 cm. 10 June 2005 ISBN 1-877407-04-6 (paperback) ISBN 1-877407-05-4 (Online edition) FIRST PUBLISHED IN 2005 BY Magnolia Press P.O. Box 41383 Auckland 1030 New Zealand e-mail: [email protected] http://www.mapress.com/zootaxa/ © 2005 Magnolia Press All rights reserved. No part of this publication may be reproduced, stored, transmitted or disseminated, in any form, or by any means, without prior written permission from the publisher, to whom all requests to reproduce copyright material should be directed in writing. This authorization does not extend to any other kind of copying, by any means, in any form, and for any purpose other than private research use. ISSN 1175-5326 (Print edition) ISSN 1175-5334 (Online edition) Zootaxa 1005: 1–64 (2005) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ ZOOTAXA 1005 Copyright © 2005 Magnolia Press ISSN 1175-5334 (online edition) The millipede type specimens in the Collections of the Field Museum of Natural History (Arthropoda: Diplopoda) PETRA SIERWALD1, JASON E. BOND2 & GRZEGORZ T. GURDA3 1Zoology, Insects, Field Museum of Natural History, 1400 S Lake Shore Drive, Chicago, Illinois 60605 2East Carolina University, Department of Biology, Howell Science complex-N211, Greenville, North Carolina 27858, USA 3 University of Michigan, Department of Molecular & Integrative Physiology 1150 W. -

The Leggiest Animal Known on Earth
The leggiest animal known on Earth With up to 750 legs, the millipede Illacme plenipes Cook and Loomis, 1928 (see photos) is the leggiest animal known on Earth. According to Marek, Shear and Bond (2012), who provided a recent, detailed redescription of the species (http://www.pensoft.net/journals/zookeys/article/3831/abstract/ ) in the periodical Zookeys, it is endemic to the northwestern foothills of the Gabilan Range in San Benito County, Silicon Valley, California. Illacme plenipes is only known from 3 localities in a 4.5 km 2 area. At present, two families are recognized in the order: Siphonophoridae and Siphonorhinidae. Among these families, three genera occur in the United States and Illacme is the only known Western Hemisphere representative of Siphonorhinidae. Illacme plenipes was described by O.F. Cook and H.F. Loomis in 1928 from seven individuals collected from a site located “a short distance after crossing the divide between Salinas and San Juan Bautista…in a small valley of a northern slope wooded with oaks, under a rather large stone”. In 2005 and 2007, new specimens were collected from near the type locality. Individuals of the species are strictly associated with large arkose sandstone boulders (see photo), and are extremely rare, with only 17 specimens known to exist in natural history collections . In contrast with its small size and unassuming outward appearance, the microanatomy of the species is strikingly complex (see photos). Based on functional morphology of related species, the extreme number of legs is hypothesized to be associated with a life spent burrowing deep underground, and clinging to the surface of sandstone boulders.