8 SI D. Chauss Et Al. Table S2 Detected EC Gene-Specific
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Sharmin Supple Legend 150706
Supplemental data Supplementary Figure 1 Generation of NPHS1-GFP iPS cells (A) TALEN activity tested in HEK 293 cells. The targeted region was PCR-amplified and cloned. Deletions in the NPHS1 locus were detected in four clones out of 10 that were sequenced. (B) PCR screening of human iPS cell homologous recombinants (C) Southern blot screening of human iPS cell homologous recombinants Supplementary Figure 2 Human glomeruli generated from NPHS1-GFP iPS cells (A) Morphological changes of GFP-positive glomeruli during differentiation in vitro. A different aggregate from the one shown in Figure 2 is presented. Lower panels: higher magnification of the areas marked by rectangles in the upper panels. Note the shape changes of the glomerulus (arrowheads). Scale bars: 500 µm. (B) Some, but not all, of the Bowman’s capsule cells were positive for nephrin (48E11 antibody: magenta) and GFP (green). Scale bars: 10 µm. Supplementary Figure 3 Histology of human podocytes generated in vitro (A) Transmission electron microscopy of the foot processes. Lower magnification of Figure 4H. Scale bars: 500 nm. (B) (C) The slit diaphragm between the foot processes. Higher magnification of the 1 regions marked by rectangles in panel A. Scale bar: 100 nm. (D) Absence of mesangial or vascular endothelial cells in the induced glomeruli. Anti-PDGFRβ and CD31 antibodies were used to detect the two lineages, respectively, and no positive signals were observed in the glomeruli. Podocytes are positive for WT1. Nuclei are counterstained with Nuclear Fast Red. Scale bars: 20 µm. Supplementary Figure 4 Cluster analysis of gene expression in various human tissues (A) Unbiased cluster analysis across various human tissues using the top 300 genes enriched in GFP-positive podocytes. -

Evolutionary Analysis of Viral Sequences in Eukaryotic Genomes
Evolutionary analysis of viral sequences in eukaryotic genomes Sean Schneider A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Washington 2014 Reading Committee: James H. Thomas, Chair Willie Swanson Phil Green Program Authorized to Offer Degree: Genome Sciences ©Copyright 2014 Sean Schneider University of Washington Abstract Evolutionary analysis of viral sequences in eukaryotic genomes Sean Schneider Chair of the supervisory committee: Professor James H. Thomas Genome Sciences The focus of this work is several evolutionary analyses of endogenous viral sequences in eukaryotic genomes. Endogenous viral sequences can provide key insights into the past forms and evolutionary history of viruses, as well as the responses of host organisms they infect. In this work I have examined viral sequences in a diverse assortment of eukaryotic hosts in order to study coevolution between hosts and the organisms that infect them. This research consisted of two major lines of investigation. In the first portion of this work, I outline the hypothesis that the C2H2 zinc finger gene family in vertebrates has evolved by birth-death evolution in response to sporadic retroviral infection. The hypothesis suggests an evolutionary model in which newly duplicated zinc finger genes are retained by selection in response to retroviral infection. This hypothesis is supported by a strong association (R2=0.67) between the number of endogenous retroviruses and the number of zinc fingers in diverse vertebrate genomes. Based on this and other evidence, the zinc finger gene family appears to act as a “genomic immune system” against retroviral infections. The other major line of investigation in this work examines endogenous virus sequences utilized by parasitic wasps to disable hosts that they infect. -

1 SUPPLEMENTAL DATA Figure S1. Poly I:C Induces IFN-Β Expression
SUPPLEMENTAL DATA Figure S1. Poly I:C induces IFN-β expression and signaling. Fibroblasts were incubated in media with or without Poly I:C for 24 h. RNA was isolated and processed for microarray analysis. Genes showing >2-fold up- or down-regulation compared to control fibroblasts were analyzed using Ingenuity Pathway Analysis Software (Red color, up-regulation; Green color, down-regulation). The transcripts with known gene identifiers (HUGO gene symbols) were entered into the Ingenuity Pathways Knowledge Base IPA 4.0. Each gene identifier mapped in the Ingenuity Pathways Knowledge Base was termed as a focus gene, which was overlaid into a global molecular network established from the information in the Ingenuity Pathways Knowledge Base. Each network contained a maximum of 35 focus genes. 1 Figure S2. The overlap of genes regulated by Poly I:C and by IFN. Bioinformatics analysis was conducted to generate a list of 2003 genes showing >2 fold up or down- regulation in fibroblasts treated with Poly I:C for 24 h. The overlap of this gene set with the 117 skin gene IFN Core Signature comprised of datasets of skin cells stimulated by IFN (Wong et al, 2012) was generated using Microsoft Excel. 2 Symbol Description polyIC 24h IFN 24h CXCL10 chemokine (C-X-C motif) ligand 10 129 7.14 CCL5 chemokine (C-C motif) ligand 5 118 1.12 CCL5 chemokine (C-C motif) ligand 5 115 1.01 OASL 2'-5'-oligoadenylate synthetase-like 83.3 9.52 CCL8 chemokine (C-C motif) ligand 8 78.5 3.25 IDO1 indoleamine 2,3-dioxygenase 1 76.3 3.5 IFI27 interferon, alpha-inducible -

The Vaccinia Virus K7 Protein Promotes Histone Methylation Associated with Heterochromatin Formation
RESEARCH ARTICLE The vaccinia virus K7 protein promotes histone methylation associated with heterochromatin formation Wondimagegnehu M. Teferi☯¤, Megan A. Desaulniers☯, Ryan S. Noyce, Mira Shenouda, Brittany Umer, David H. Evans* Department of Medical Microbiology & Immunology, Li Ka Shing Institute of Virology, University of Alberta, Edmonton, Alberta, Canada ☯ These authors contributed equally to this work. ¤ Current address: Department of Medicine, Jacobi Medical Center, Albert Einstein College of Medicine, a1111111111 Bronx, New York, United States of America a1111111111 * [email protected] a1111111111 a1111111111 a1111111111 Abstract It has been well established that many vaccinia virus proteins suppress host antiviral path- ways by targeting the transcription of antiviral proteins, thus evading the host innate immune system. However, whether viral proteins have an effect on the host's overall cellular tran- OPEN ACCESS scription is less understood. In this study we investigated the regulation of heterochromatin Citation: Teferi WM, Desaulniers MA, Noyce RS, during vaccinia virus infection. Heterochromatin is a highly condensed form of chromatin Shenouda M, Umer B, Evans DH (2017) The vaccinia virus K7 protein promotes histone that is less transcriptionally active and characterized by methylation of histone proteins. We methylation associated with heterochromatin examined the change in methylation of two histone proteins, H3 and H4, which are major formation. PLoS ONE 12(3): e0173056. markers of heterochromatin, during the course of viral infection. Using immunofluorescence doi:10.1371/journal.pone.0173056 microscopy and flow cytometry we were able to track the overall change in the methylated Editor: Janet F. Partridge, Saint Jude Children's levels of H3K9 and H4K20. Our results suggest that there is significant increase in methyla- Research Hospital, UNITED STATES tion of H3K9 and H4K20 during Orthopoxviruses infection compared to mock-infected cells. -

Nucleus Accumbens-Associated Protein 1 Binds DNA Directly Through the BEN Domain in a Sequence-Specific Manner
biomedicines Article Nucleus Accumbens-Associated Protein 1 Binds DNA Directly through the BEN Domain in a Sequence-Specific Manner Naomi Nakayama 1,2, Gyosuke Sakashita 1, Takashi Nagata 3,4 , Naohiro Kobayashi 5, Hisashi Yoshida 6, Sam-Yong Park 6, Yuko Nariai 1, Hiroaki Kato 1, Eiji Obayashi 1, Kentaro Nakayama 7, Satoru Kyo 7 and Takeshi Urano 1,* 1 Department of Biochemistry, Shimane University School of Medicine, Izumo, Shimane 693-8501, Japan; [email protected] (N.N.); [email protected] (G.S.); [email protected] (Y.N.); [email protected] (H.K.); [email protected] (E.O.) 2 Department of Health and Nutrition, The University of Shimane, Izumo, Shimane 693-8550, Japan 3 Institute of Advanced Energy, Kyoto University, Kyoto 606-8501, Japan; [email protected] 4 Graduate School of Energy Science, Kyoto University, Kyoto 606-8501, Japan 5 Institute for Protein Research, Osaka University, Suita, Osaka 565-0871, Japan; [email protected] 6 Protein Design Laboratory, Graduate School of Medical Life Science, Yokohama City University, Yokohama, Kanagawa 230-0045, Japan; [email protected] (H.Y.); [email protected] (S.-Y.P.) 7 Department of Obstetrics and Gynecology, Shimane University School of Medicine, Izumo, Shimane 693-8501, Japan; [email protected] (K.N.); [email protected] (S.K.) * Correspondence: [email protected]; Tel.: +81-853-20-2126 Received: 2 December 2020; Accepted: 10 December 2020; Published: 14 December 2020 Abstract: Nucleus accumbens-associated protein 1 (NAC1) is a nuclear protein that harbors an amino-terminal BTB domain and a carboxyl-terminal BEN domain. -

Novel Noncatalytic Substrate-Selective P38α-Specific
Novel Noncatalytic Substrate-Selective p38α -Specific MAPK Inhibitors with Endothelial-Stabilizing and Anti-Inflammatory Activity This information is current as of September 26, 2021. Nirav G. Shah, Mohan E. Tulapurkar, Aparna Ramarathnam, Amanda Brophy, Ramon Martinez III, Kellie Hom, Theresa Hodges, Ramin Samadani, Ishwar S. Singh, Alexander D. MacKerell, Jr., Paul Shapiro and Jeffrey D. Hasday J Immunol 2017; 198:3296-3306; Prepublished online 15 Downloaded from March 2017; doi: 10.4049/jimmunol.1602059 http://www.jimmunol.org/content/198/8/3296 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2017/03/15/jimmunol.160205 Material 9.DCSupplemental References This article cites 61 articles, 17 of which you can access for free at: http://www.jimmunol.org/content/198/8/3296.full#ref-list-1 by guest on September 26, 2021 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2017 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Novel Noncatalytic Substrate-Selective p38a-Specific MAPK Inhibitors with Endothelial-Stabilizing and Anti-Inflammatory Activity Nirav G. -

A Novel 80-Nm Virus Infecting Acanthamoeba Castellanii
Yaravirus: A novel 80-nm virus infecting Acanthamoeba castellanii Paulo V. M. Borattoa,b,1, Graziele P. Oliveiraa,b,1, Talita B. Machadoa, Ana Cláudia S. P. Andradea, Jean-Pierre Baudoinb,c, Thomas Klosed, Frederik Schulze, Saïd Azzab,c, Philippe Decloquementb,c, Eric Chabrièreb,c, Philippe Colsonb,c, Anthony Levasseurb,c, Bernard La Scolab,c,2, and Jônatas S. Abrahãoa,2 aLaboratório de Vírus, Instituto de Ciências Biológicas, Departamento de Microbiologia, Universidade Federal de Minas Gerais, Belo Horizonte, MG 31270-901, Brazil; bMicrobes, Evolution, Phylogeny and Infection, Aix-Marseille Université UM63, Institut de Recherche pour le Développement 198, Assistance Publique–Hôpitaux de Marseille, 13005 Marseille, France; cInstitut Hospitalo-Universitaire Méditerranée Infection, Faculté de Médecine, 13005 Marseille, France; dDepartment of Biological Sciences, Purdue University, West Lafayette, IN 47907; and eDepartment of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720 Edited by James L. Van Etten, University of Nebraska-Lincoln, Lincoln, NE, and approved June 2, 2020 (received for review January 29, 2020) Here we report the discovery of Yaravirus, a lineage of amoebal others. NCDLVs have dsDNA genomes and were proposed to virus with a puzzling origin and evolution. Yaravirus presents share a monophyletic origin based on criteria that include the 80-nm-sized particles and a 44,924-bp dsDNA genome encoding sharing of a set of ancestral vertically inherited genes (17, 18). for 74 predicted proteins. Yaravirus genome annotation showed From this handful set of genes, a core gene cluster is found to be that none of its genes matched with sequences of known organ- present in almost all members of the NCLDVs, being composed isms at the nucleotide level; at the amino acid level, six predicted by five distinct genes, namely a DNA polymerase family B, a proteins had distant matches in the nr database. -

Individual Functions and Substrate Specificities of Importin Alpha
From the Max-Delbrück-Center for Molecular Medicine Berlin Head of the research group: Prof. Dr. M. Bader “Individual functions and substrate specificities of importin α subtypes” Dissertation for Fulfillment of Requirements for the Doctoral Degree of the University of Lübeck from the Department of Natural Sciences Submitted by Stefanie Hügel from Hennigsdorf Lübeck 2013 First referee: Prof. Dr. M. Bader Second referee: Prof. Dr. N. Tautz Date of oral examination: 21.02.13 Approved for printing. Lübeck, 22.02.2013 2 1 TABLE OF CONTENTS 1 TABLE OF CONTENTS _______________________________________________________ 3 2 ABSTRACT _______________________________________________________________ 6 3 ZUSAMMENFASSUNG ______________________________________________________ 7 4 INTRODUCTION ___________________________________________________________ 9 4.1 Nucleocytoplasmic protein transport ____________________________________________ 9 4.1.1 The nuclear pore complex – gateway to the nucleus ______________________________________ 10 4.1.1 Transport factors __________________________________________________________________ 11 4.1.2 Nothing happens without RanGTP _____________________________________________________ 12 4.2 The classical nuclear protein import pathway ____________________________________ 13 4.2.1 Structure and function of importin α ___________________________________________________ 13 4.2.2 Importin α:β mediated nuclear protein import___________________________________________ 15 4.2.3 The importin α protein family ________________________________________________________ -

Dinshaw J. PATEL
Dinshaw J. PATEL (cv) (updated 12-29-16) Member and Abby Rockefeller Mauzé Chair in Experimental Therapeutics Structural Biology Program Memorial Sloan-Kettering Cancer Center 1275 York Avenue New York, NY, 10065, USA Phone (office): 1-212-639-7207 Phone (cell): 1-914-980-0896 FAX: 1-212-717-3066 Email: [email protected] Web site: http://www.mskcc.org/mskcc/html/10829.cfm Date and Place of Birth: April 25, 1942; Mumbai, India. Naturalized US citizen (1990). Education 1961 B.Sc. University of Mumbai, Mumbai, India. Chemistry 1963 M.S. California Institute of Technology, Pasadena, CA. Chemistry 1968 PhD. New York University, New York, NY. Chemistry Postdoctoral Training 1967 New York Univ. Medical School New York, NY. Biochemistry. 1968 - 1969 AT&T Bell Laboratories, Murray Hill, NJ. Biophysics. Appointments 1970 - 1984 Member of Technical Staff, Polymer Chemistry Department, AT&T Bell Laboratories, Murray Hill, NJ 1984 - 1992 Professor of Biochemistry & Molecular Biophysics, College of Physicians & Surgeons, Columbia University, New York, NY 1992 - Member, Structural Biology Program Memorial Sloan-Kettering Cancer Center (MSKCC), New York, New York 1994 - Professor, Graduate Program in Biochemistry & Structural Biology, Weill School of Medical Sciences, Cornell University, New York, NY Honors and Awards 1961 - 1963 Jamshetjee N. Tata Fellow 1983 AT&T Bell Laboratories Distinguished Technical Staff Award 1992 - Abby Rockefeller Mauzé Chair in Experimental Therapeutics (MSKCC) 1997 Distinguished Alumnus Award, New York University 1997 - 1999 Harvey Society (Vice-President 97-98; President 98-99) 2013 NIH Directors Transformative R01 Award (with Thomas Tuschl and Uwe Ohler) 2014 2014 FEZANA Jamshed and Shirin Guzdar Excellence in Profession Award Academy Memberships 2009 Member, National Academy of Sciences, USA 2014 Member, American Academy of Arts and Sciences, USA Named/Plenary/Keynote Lectureships (since 2010) 2010 John D. -
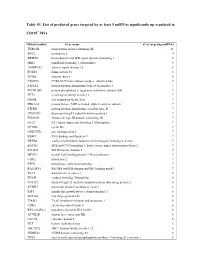
Table S3. List of Predicted Genes Targeted by at Least 5 Mirnas Significantly up Regulated In
Table S3. List of predicted genes targeted by at least 5 miRNAs significantly up regulated in CD105+ MVs. Official symbol Gene name n° of targeting miRNAs TNRC6B trinucleotide repeat containing 6B 11 BNC2 basonuclin 2 9 BRWD1 bromodomain and WD repeat domain containing 1 8 MIB1 mindbomb homolog 1 (Drosophila) 8 ANKRD52 ankyrin repeat domain 52 8 FOXP1 forkhead box P1 8 ITGB8 integrin, beta 8 8 CNOT6L CCR4-NOT transcription complex, subunit 6-like 8 PTP4A1 protein tyrosine phosphatase type IVA, member 1 7 PPP1R15B protein phosphatase 1, regulatory (inhibitor) subunit 15B 7 TET3 tet oncogene family member 3 7 GMFB glia maturation factor, beta 7 PRKAA2 protein kinase, AMP-activated, alpha 2 catalytic subunit 7 PTPRD protein tyrosine phosphatase, receptor type, D 7 TP53INP1 tumor protein p53 inducible nuclear protein 1 7 FNDC3B fibronectin type III domain containing 3B 7 FAT3 FAT tumor suppressor homolog 3 (Drosophila) 7 CCND2 cyclin D2 7 ONECUT2 one cut homeobox 2 7 RBM9 RNA binding motif protein 9 7 ERBB4 v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) 7 BACH2 BTB and CNC homology 1, basic leucine zipper transcription factor 2 7 SMAD4 SMAD family member 4 7 MECP2 methyl CpG binding protein 2 (Rett syndrome) 7 UBN2 ubinuclein 2 7 PTEN phosphatase and tensin homolog 7 RALGPS1 Ral GEF with PH domain and SH3 binding motif 1 7 DLC1 deleted in liver cancer 1 6 ENAH enabled homolog (Drosophila) 6 NUFIP2 nuclear fragile X mental retardation protein interacting protein 2 6 KCMF1 potassium channel modulatory factor 1 6 IGF1 insulin-like growth factor 1 (somatomedin C) 6 ZNF148 zinc finger protein 148 6 TIAM1 T-cell lymphoma invasion and metastasis 1 6 CDK6 cyclin-dependent kinase 6 6 RP5-1022P6.2 hypothetical protein KIAA1434 6 ACVR2B activin A receptor, type IIB 6 CLCN5 chloride channel 5 6 HLF hepatic leukemia factor 6 TBC1D12 TBC1 domain family, member 12 6 FRMD4A FERM domain containing 4A 6 NUS1 nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. -
Supplemental Table 1 Genes Up-Regulated (>2.0) in Stat3
Supplemental Table 1 Genes up-regulated (>2.0) in Stat3∆/∆ compared with Stat3flox/flox Probe ID Gene Symbol Gene Description Entrez gene ID 1460470_at Acoxl acyl-Coenzyme A oxidase-like 74121 1459876_at --- --- --- 1459875_x_at 5730494M16Rik RIKEN cDNA 5730494M16 gene 66648 1459841_x_at Laptm5 lysosomal-associated protein transmembrane 5 16792 1459118_at D230038C21 hypothetical protein D230038C21 328872 1459018_at D10Ertd447e DNA segment, Chr 10, ERATO Doi 447, expressed 52607 1458924_at D430013B06Rik RIKEN cDNA D430013B06 gene 399594 1458729_at --- --- --- 1458712_at --- --- --- 1458274_at Zfp69 zinc finger protein 69 381549 1457966_at --- --- --- 1457851_at --- --- --- 1457743_at --- --- --- 1457633_x_at Cox6a2 cytochrome c oxidase, subunit VI a, polypeptide 2 12862 1457589_at Fat3 FAT tumor suppressor homolog 3 (Drosophila) 270120 1457491_at Plekha1 Pleckstrin homology domain containing, family A (phosphoinositide binding 101476 specific) member 1 1457332_at Prmt3 protein arginine N-methyltransferase 3 71974 1457247_at Lmln leishmanolysin-like (metallopeptidase M8 family) 239833 1457129_at LOC100504796 hypothetical LOC100504796 100504796 1456854_at Neurl1a neuralized homolog 1A (Drosophila) 18011 1456794_at E230016M11Rik RIKEN cDNA E230016M11 gene 320172 1456793_at Cytl1 cytokine-like 1 231162 1456780_at --- --- --- 1456527_at Hecw1 HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 94253 1456418_at Kcnj13 potassium inwardly-rectifying channel, subfamily J, member 13 100040591 1456404_at Adamts5 a disintegrin-like and