Towards a Phylogenetic Framework for the Evolution of Shakes, Rattles, and Rolls in Myiarchus Tyrant-flycatchers (Aves: Passeriformes: Tyrannidae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Birds of Reserva Ecológica Guapiaçu (REGUA)
Cotinga 33 The birds of Reserva Ecológica Guapiaçu (REGUA), Rio de Janeiro, Brazil Leonardo Pimentel and Fábio Olmos Received 30 September 2009; final revision accepted 15 December 2010 Cotinga 33 (2011): OL 8–24 published online 16 March 2011 É apresentada uma lista da avifauna da Reserva Ecológica de Guapiaçu (REGUA), uma reserva privada de 6.500 ha localizada no município de Cachoeiras de Macacu, vizinha ao Parque Estadual dos Três Picos, Estação Ecológica do Paraíso e Parque Nacional da Serra dos Órgãos, parte de um dos maiores conjuntos protegidos do Estado do Rio de Janeiro. Foram registradas um total de 450 espécies de aves, das quais 63 consideradas de interesse para conservação, como Leucopternis lacernulatus, Harpyhaliaetus coronatus, Triclaria malachitacea, Myrmotherula minor, Dacnis nigripes, Sporophila frontalis e S. falcirostris. A reserva também está desenvolvendo um projeto de reintrodução dos localmente extintos Crax blumembachii e Aburria jacutinga, e de reforço das populações locais de Tinamus solitarius. The Atlantic Forest of eastern Brazil and Some information has been published on neighbouring Argentina and Paraguay is among the birds of lower (90–500 m) elevations in the the most imperilled biomes in the world. At region10,13, but few areas have been subject to least 188 bird species are endemic to it, and 70 long-term surveys. Here we present the cumulative globally threatened birds occur there, most of them list of a privately protected area, Reserva Ecológica endemics4,8. The Atlantic Forest is not homogeneous Guapiaçu (REGUA), which includes both low-lying and both latitudinal and longitudinal gradients parts of the Serra dos Órgãos massif and nearby account for diverse associations of discrete habitats higher ground, now mostly incorporated within and associated bird communities. -

CAT Vertebradosgt CDC CECON USAC 2019
Catálogo de Autoridades Taxonómicas de vertebrados de Guatemala CDC-CECON-USAC 2019 Centro de Datos para la Conservación (CDC) Centro de Estudios Conservacionistas (Cecon) Facultad de Ciencias Químicas y Farmacia Universidad de San Carlos de Guatemala Este documento fue elaborado por el Centro de Datos para la Conservación (CDC) del Centro de Estudios Conservacionistas (Cecon) de la Facultad de Ciencias Químicas y Farmacia de la Universidad de San Carlos de Guatemala. Guatemala, 2019 Textos y edición: Manolo J. García. Zoólogo CDC Primera edición, 2019 Centro de Estudios Conservacionistas (Cecon) de la Facultad de Ciencias Químicas y Farmacia de la Universidad de San Carlos de Guatemala ISBN: 978-9929-570-19-1 Cita sugerida: Centro de Estudios Conservacionistas [Cecon]. (2019). Catálogo de autoridades taxonómicas de vertebrados de Guatemala (Documento técnico). Guatemala: Centro de Datos para la Conservación [CDC], Centro de Estudios Conservacionistas [Cecon], Facultad de Ciencias Químicas y Farmacia, Universidad de San Carlos de Guatemala [Usac]. Índice 1. Presentación ............................................................................................ 4 2. Directrices generales para uso del CAT .............................................. 5 2.1 El grupo objetivo ..................................................................... 5 2.2 Categorías taxonómicas ......................................................... 5 2.3 Nombre de autoridades .......................................................... 5 2.4 Estatus taxonómico -

The Lesser Antilles Incuding Trinidad
The brilliant Lesser Antillean Barn Owl again showed superbly. One of several potential splits not yet recognized by the IOC (Pete Morris) THE LESSER ANTILLES INCUDING TRINIDAD 5 – 20/25 JUNE 2015 LEADERS: PETE MORRIS After our successful tour around the Caribbean in 2013, it was great to get back again this year. It all seemed pretty straightforward this time around, and once again we cleaned up on all of the available endemics, po- 1 BirdQuest Tour Report:The Lesser Antilles www.birdquest-tours.com The fabulous White-breasted Thrasher from Martinique (Pete Morris) tential splits and other goodies. For sure, this was no ordinary Caribbean holiday! During the first couple of weeks we visited no fewer than ten islands (Antigua, Barbuda, Montserrat, Dominica, Guadeloupe, Martinique, St Lucia, St Vincent, Barbados and Grenada), a logistical feat of some magnitude. With plenty of LIAT flights (the islanders refer to LIAT as ‘Leave Island any Time’ and ‘Luggage in Another Terminal’ to name but two of the many funny phrases coined from LIAT) and unreliable AVIS car hire reservations, we had our work cut out, but in the end, all worked out! It’s always strange birding on islands with so few targets, but with so many islands to pack-in, we were never really short of things to do. All of the endemics showed well and there were some cracking highlights, including the four smart endemic amazons, the rare Grenada Dove, the superb Lesser Antillean Barn Owl, the unique tremblers and White-breasted Thrashers, and a series of colourful endemic orioles to name just a few! At the end of the Lesser Antilles adventure we enjoyed a few days on Trinidad. -

Pipridae) and of the Cotingas (Cotingidae) Based on Morphology
OCCASIONAL PAPERS OF THE MUSEUM OF ZOOLOGY THE UNIVERSITY OF MICHIGAN A TEST OF THE MONOPHYLY OF THE MANAKINS (PIPRIDAE) AND OF THE COTINGAS (COTINGIDAE) BASED ON MORPHOLOGY ABSTRACT.-Pmm, Richard 0. A test of the monophyly of the manakins (Pipridae) and of the cotingas (Cotingidae) based on morphology. Occ. Pap. Mus. Zool., Uniu. Michigan, 723:I-44,6jigs. A phylogenetic analysis of the Tyr- annoidea is performed as a test of the monophyly of the manakins (Pipri- dae) and of the cotingas (Cotingidae). The 12 morphological characters surveyed include the traditional characters used to define the families and other morphological features taken from observations of tyrannoid syr- inges and hindlimb arteries. Five traditional characters are phylogeneti- cally uninformative. The remaining seven characters support 25 maxi- mally parsimonious phylogenetic hypotheses of length 10 (CI = 0.70). A strict consensus tree based on these trees has few resolved clades, but indicates that neither the Pipridae nor the Cotingidae as traditionally defined is monophyletic. Six currently recognized genera of Pipridae- Schiffornis, Sapayoa, Piprites, Neopipo, Neopelma, and Tyranneutes-share de- rived morphological characters with other, non-piprid tyrannoids. The other eleven piprid genera-4hloropip0, Xenopipo, Antilophia, Heterocercus, Machaeropterus, Manacus, Corapipo, Ilicura, Masiur, Chiroxiphia, and Pipra- form a clade diagnosed by the dorsal fusion of the B1-2 syringeal sup- porting elements. A large clade including most cotingids is supported by a derived syringeal muscle character and provides evidence of the mono- phyly of the cotingids, but this character conflicts with other derived morphological features. Additional data are required to resolve many portions of tyrannoid higher-level phylogeny. -

Reference File
References added since publication of 2007 CRC Handbook of Avian Body Masses Abadie, K. B., J. Pérez Z., and M. Valverde. 2006. Primer reporte de colonias del Martín Peruano Progne murphyi. Cotinga 24:99-101. Ackerman, J. T., J. Y. Takekawa, J. D. Bluso, J. L. Yee, and C. A. Eagles-Smith. 2008. Gender identification of Caspian Terns using external morphology and discriminant function analysis. Wilson Journal of Ornithology 120:378-383. Alarcos, S., C. de la Cruz, E. Solís, J. Valencia, and M. J. García-Baquero. 2007. Sex determination of Iberian Azure-winged Magpies Cyanopica cyanus cooki by discriminant analysis of external measurements. Ringing & Migration 23:211-216. Albayrak, T., A. Besnard, and A. Erdoğan. 2011. Morphometric variation and population relationships of Krüeper’s Nuthatch (Sitta krueperi) in Turkey. Wilson Journal of Ornithology 123:734-740. Aleixo, A., C. E. B. Portes, A. Whittaker, J. D. Weckstein, L. Pedreira Gonzaga, K. J. Zimmer, C. C. Ribas, and J. M. Bates. 2013. Molecular systematics and taxonomic revision of the Curve-billed Scythebill complex (Campylorhamphus procurvoides: Dendrocolaptidae), with description of a new species from western Amazonian Brazil. Pp. 253-257, In: del Hoyo, J., A Elliott, J. Sargatal, and D.A. Christie (eds). Handbook of the birds of the world. Special volume: new species and global index. Lynx Edicions, Barcelona, Spain. Volume 1. Alfano, A. 2014. Pygmy Nightjar (Nyctopolus hirundinaeus). Neotropical Birds Online (T.S. Schulenberg, ed.). Cornell Laboratory of Ornithology, Ithaca, NY. Alvarenga, H. M. F., E. Höfling, and L. F. Silveira. 2002. Notharchus swainsoni (Gray, 1846) é uma espécie válida. -

Avian Monitoring Program
AVIAN INVENTORY AND MONITORING REPORT OSA CONSERVATION PROPERTIES CERRO OSA PIRO NEENAH PAPER OSA PENINSULA, COSTA RICA PREPARED BY: KAREN M. LEAVELLE FOR: OSA CONSERVATION APRIL 2013 Scarlet Macaw © Alan Dahl TABLE OF CONTENTS INTRODUCTION 2 METHODS 3 Study Site 3 Bird Surveys 5 RESULTS Community Composition and Density 6 Neotropical Migratory and Indicator Species 6 Habitat and Elevation Associations Neenah Paper 12 LITERATURE CITED 14 Table 1: Osa Priority Species 3 Table 2: Species Richness 6 Table 3: Cumulative list of resident bird species 7 Table 4: Cumulative list of Neotropical migratory birds 10 Table 5: BCAT by elevation 11 Table 6: BCAT by forest type 12 Table 7: Neenah Paper species richness 12 Appendix A: Bird species densities Osa Conservation 15 Appendix B: Bird species densities Neenah Paper 16 Appendix C: Threatened or endemic species 17 Appendix D: Comprehensive list of all OC bird species 18 Appendix E: Comprehensive list of all Osa Peninsula species 24 RECOMMENDED CITATION Leavelle, K.M. 2013. Avian Inventory and Monitoring Report for Osa Conservation Properties at Cerro Osa and Piro Research Stations, Osa Peninsula, Costa Rica. Technical Report for Osa Conservation. p 36. Washington, DC. INTRODUCTION The Osa Peninsula of Costa Rica is home to over 460 tropical year round resident and overwintering neotropical migratory bird species blanketing one of the most biologically diverse corners of the planet. The Osa habors eight regional endemic species, five of which are considered to be globally threatened or endangered (Appendix C), and over 100 North American Nearctic or passage migrants found within all 13 ecosystems that characterize the peninsula. -
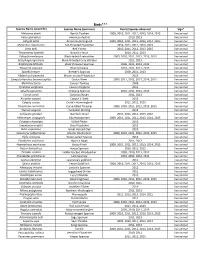
Flora and Fauna List.Xlsx
Birds*** Species Name (scientific) Species Name (common) Year(s) Species observed Sign* Melozone aberti Abert's Towhee 2009, 2010, 2011, 2012, 2013, 2014, 2015 live animal Falco sparverius American Kestrel 2010, 2013 live animal Calypte anna Anna's Hummingbird 2009, 2010, 2011, 2012, 2013, 2014, 2015 live animal Myiarchus cinerascens Ash‐throated Flycatcher 2010, 2011, 2012, 2013, 2015 live animal Vireo bellii Bell's Vireo 2010, 2011, 2012, 2013, 2015 live animal Thryomanes bewickii Bewick's Wren 2010, 2011, 2013 live animal Polioptila melanura Black‐tailed Gnatcatcher 2009, 2010, 2011, 2012, 2013, 2015 live animal Setophaga nigrescens Black‐throated Gray Warbler 2011, 2013 live animal Amphispiza bilineata Black‐throated Sparrow 2009, 2011, 2012, 2013 live animal Passerina caerulea Blue Grosbeak 2010, 2011, 2012, 2013 live animal Spizella breweri Brewer's Sparrow 2009, 2011, 2013 live animal Myiarchus tryannulus Brown‐crested Flycatcher 2013 live animal Campylorhynchus brunneicapillus Cactus Wren 2009, 2011, 2012, 2013, 2014, 2015 live animal Melozone fusca Canyon Towhee 2009 live animal Tyrannus vociferans Cassin's Kingbird 2011 live animal Spizella passerina Chipping Sparrow 2010, 2011, 2012, 2013 live animal Corvus corax Common Raven 2011, 2013 live animal Accipiter cooperii Cooper's Hawk 2013 live animal Calypte costae Costa's Hummingbird 2011, 2012, 2013 live animal Toxostoma curvirostre Curve‐billed Thrasher 2009, 2010, 2011, 2012, 2013, 2015 live animal Sturnus vulgaris European Starling 2012 live animal Callipepla gambelii Gambel's -

20100512 Myiarchus Flycatchers P304
Notes based on Joe Morlan’s Ornithology class lecture May 12 th , 2010. Joe Morlan is not responsible for these notes, any errors or omissions in them are mine. You can find Western Tanagers in flocks during migration. There have been occasional reports of large numbers of migrants very locally on certain ridges these last few weeks. Also a lot of reports of people seeing almost no migrants. It seems that migration is much more localized this year, not widespread. Some of the birds looked for during the class field trip to Briones did not show up. The wet winter may be a reason. Birds of arid valleys and deserts are not forced to go away from the dry interior, they may have enough water there to be able to nest locally instead of being pushed far. When there are invasions of species like Black-throated Sparrows there was usually a wet year followed by a draught. They have excellent breeding success during the wet year and then there are too many birds during the draught, they get pushed into areas where they would not normally be. Bird nest boxes come in different sizes and with different hole sizes that have been specified for particular birds. Also the height at which you put up the box can affect which species you get. Starlings do not like to nest in bird houses that are below six or seven feet from the ground. Bluebirds are well studied nest box users because of the big decline in bluebirds due to starlings occupying old woodpecker holes. -

Lapa Rios Bird Checklist Lapa Rios Bird Checklist
Lapa Rios Bird Checklist Lapa Rios Bird Checklist The birds listed as "have been seen" at Lapa Rios include the Reserve itself as well as sighthings in the Matapalo (beach) area, in and around Puerto Jiménez and along the road from Puerto Jiménez to Lapa Rios; a distance of approximately 19 kilometers (11 miles). Lapa Rios is a private Biological Preserve of approximately 1000 acres. Access to its trail system is only through the permission of the management. The trail inmmediately adjacent to the main lodge can be explored without a staff guide, but a staff guide is required for any excursion into the interior of the preserve or along the Carbonera River. STATUS CODE: A = "Abundant" - many seen or heard daily in appropriate habitat/season and/or in large groups at frequent intervals. C = "Common" - consistently recorded in appropiate habitat/season and/or in large groups at frequent intervals. U = "Uncommon" - recorded regularly but with longer intervals and in small numbers. R = "Rare" - recorded in very small numbers or on really rare occasions. Acc = "Accidental" - recorded only a few times at Lapa Rios sometimes far out of its normal range and not likely to recur. Ex = "Extinct"- considered to be extint in the wild, with no populations on the country and only few sightings in the last years. GARRIGUES GUIDE: We reference Richard Garrigues guidebook for the bird’s description. The Birds of Costa Rica: A Field Guide. Zona Tropical Publications, Paperback – April 12, 2007 1 COMMON NAME LATIN NAME STATUS GUIDE TINAMOUS 1 Great Tinamou Tinamus major A Pag. -

Check List 5(2): 222–237, 2009
Check List 5(2): 222–237, 2009. ISSN: 1809-127X LISTS OF SPECIES Birds (Aves), Serrania Sadiri, Parque Nacional Madidi, Depto. La Paz, Bolivia Peter Andrew Hosner 1 Kenneth David Behrens 2 A. Bennett Hennessey 3 1 University of Kansas, Museum of Natural History, Ecology and Evolutionary Biology, Division of Ornithology. Dyche Hall, 1345 Jayhawk Blvd., University of Kansas, Lawrence, KS 66046. E-mail: [email protected] 2 Tropical Birding, 1 Toucan Way. Bloubergrise 7441, South Africa. 3 Asociación Civil Armonía. Avenida Lomas de Arena, Casilla 3566, Santa Cruz, Bolivia. Abstract We surveyed the Serrania Sadiri for birds at elevations between 500-950m for a combined total of 15 days in three different months. The area surveyed was along the Tumupasa/San Jose de Uchupiamones trail at the edge of Parque Nacional Madidi in Depto. La Paz, Bolivia. We report observations of 231 species of birds detected by sight and sound, including many outlying ridge specialists. We report and present photographs of a new species for Depto. La Paz (Caprimulgis nigrescens), the second Bolivian localities for Porphyrolaema prophyrolaema, Zimerius cinereicapillus, and Basileuterus chrysogaster, and five new species records for Parque Nacional Madidi. Introduction Foothills and outlying ridges of the Andes are From the small village of Tumupasa (14°8'46" S, often very difficult or impossible to access. As a 67°53'17" W; 400 m a.s.l; Figures 1 and 2), an old result, many of the specialist bird species in these trail leads generally southwest over the Serrania areas are poorly known and some only recently Sadiri to the town of San Jose de Uchupiamones described, and these areas generally have unique (14°12'47" S, 68°03'14" W; 520 m a.s.l). -

Notes and News
24 PAGEET AL. [Auk, Vol. 100 Bird Observatory. Ralph Widrig, Steven Cellman, tain Plover social systems.Living Bird 12: 69- Barbara Swarth, Susan Peaslee, Erica Buhrman, Don- 94. na Vaiano, Paul Neal, Sharon Goldwasser, Dennis HAYS, H., & M. LECI•o¾. 1971. Field criteria for Parker, John Harris, David Shuford, Stuart Johnson, determining incubation stage of the Common John and Ricky Warriner, Stephanie Zeiler, Elliot Tern. Wilson Bull. 83: 425-429. Burch, Brett Engstrom,Leslie Smith, BernadetteAl- HOLMES,R.T. 1966. Breeding ecologyand annual len, Greg Calllet, Tom Pogson,Tom Cassidy, Geoff cycle adaptationsof the Red-backedSandpiper Geupel, Doug Burgess,Judy Wagner, Kim Sullivan, (Calidrisalpina) in northern Alaska. Condor 68: JanetKjelmyr, and Tad Theimerhelped with the field 3-46. work and Philip Henderson, Carolyn Frederiksen, 1972. Ecologicalfactors influencing the and Frances Bidstrup provided valuable observa- breeding season schedule of Western Sandpi- tions. John and Ricky Warriner generouslyshared pers (Calidrisrnauri) in subarcticAlaska. Amer. with us their knowledge of Snowy Plovers at Mon- Midi. Natur. 87: 472-491. terey Bay, and CharlesSimis provided weather data. MAYFIELr•,H. F. 1961. Nesting successcalculated Jan Simis allowed us to camp on her land and ex- from exposure.Wilson Bull. 73: 255-261. tended to us very warm hospitality while we were --. 1975. Suggestionsfor calculatingnest suc- there. David Ainley, Peter Connors,David DeSante, cess. Wilson Bull. 87: 456-466. Marshall Howe, Harriet Huber, Frank Pitelka, and OI•INO, L. W., & M. L. KNOrrSON. 1972. Monogamy Chris Ribic provided helpful comments on the and polyandry in the SpottedSandpiper. Living manuscript.We thank theseorganizations and peo- Bird 11: 59-72. -

Western Mexico
Cotinga 14 W estern Mexico: a significant centre of avian endem ism and challenge for conservation action A. Townsend Peterson and Adolfo G. Navarro-Sigüenza Cotinga 14 (2000): 42–46 El endemismo de aves en México está concentrado en el oeste del país, pues entre el 40 al 47% de las aves endémicas de México están totalmente restringidas a la región. Presentamos un compendio de estos taxones, tanto siguiendo el concepto biológico de especie como el concepto filogenético de especie, documentando la región como un importante centro de endemismo. Discutimos estrategias de conservación en la región, especialmente la idea de ligar reservas para preservar transectos altitudinales de hábitats continuos, desde las tierras bajas hasta las mayores altitudes, en áreas críticas. Introduction and Transvolcanic Belt of central and western Mexico has been identified as a megadiverse coun Mexico were identified as major concentrations of try, with impressive diversity in many taxonomic endemic species. This non-coincidence of diversity groups20. Efforts to document the country’s biologi and endemism in Mexican biodiversity has since cal diversity are at varying stages of development been documented on different spatial scales13,17 and in different taxa17,19,20 but avian studies have ben in additional taxonomic groups17. efited from extensive data already accumulated18 In prior examinations, however, western Mexico and have been able to advance to more detailed lev (herein defined as the region from Sonora and Chi els of analysis6,12,17. huahua south to Oaxaca, including the coastal In the only recent countrywide survey of avian lowlands, the Sierra Madre Occidental and Sierra diversity and endemism6, the south-east lowlands Madre del Sur, and Pacific-draining interior basins were identified as important foci of avian species such as the Balsas Basin) has not been appreciated richness.