Target Selection and Annotation for the Structural Genomics of the Amidohydrolase and Enolase Superfamilies
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Dr. Martin St. Maurice's Publications
Dr. Martin St. Maurice’s Publications 2013 Lin, Y., and St. Maurice, M. 2013. The structure of allophanate hydrolase from Granulibacter bethesdensis provides insights into substrate specificity in the amidase signature family. Biochemistry. 52: 690-700. 2012 Waldrop, G.L., Holden, H.M., and St. Maurice, M. 2012. The enzymes of biotin dependent CO2 metabolism: What structures reveal about their reaction mechanisms. Protein Science 21(11):1597-1619. Adina-Zada, A., Sereeruk, C., Jitrapakdee, S., Zeczycki, T.N., St. Maurice, M., Cleland, W.W., Wallace, J.C., and Attwood, P.V. 2012. Roles of Arg427 and Arg472 in the binding and allosteric effects of acetyl CoA in pyruvate carboxylase. Biochemistry 51(41): 1597-1619. 2011 Adina-Zada, A., Hazra, R., Sereeruk, C., Jitrapakdee, S., Zeczycki, T.N., St. Maurice, M., Cleland, W.W., Wallace, J.C., and Attwood, P.V. 2011. Probing the allosteric activation of pyruvate carboxylase using 2′,3′-O-(2,4,6-trinitrophenyl) adenosine 5′-triphosphate as a fluorescent mimic of the allosteric activator acetyl CoA. Arch. Biochem. Biophys. 117-126. Zeczycki, T.N., Menefee, A.L., Jitrapakdee, S., Wallace, J.C., Attwood, P.V., St. Maurice, M. and Cleland, W.W. 2011. Activation and inhibition of pyruvate carboxylase from Rhizobium etli. Biochemistry. 9694-9707. Lietzan, A.D., Menefee, A.L., Zeczycki, T.N., Kumar, S., Attwood, P.V., Wallace, J.C., Cleland, W.W. and St. Maurice, M. 2011. Interaction between the biotin carrier domain and the biotin carboxylase domain in the structure of Rhizobium etli pyruvate carboxylase. Biochemistry. 9708-9723. Zeczycki, T.N., Menefee, A.L., Adina-Zada, A., Surinya, K.H., Wallace, J.C., Attwood, P.V., St. -

Comparison of Topological Clustering Within Protein Networks Using Edge
Comparison of topological clustering within protein networks using edge metrics that evaluate full sequence, full structure, and active site microenvironment similarity Janelle B. Leuthaeuser,1 Stacy T. Knutson,2 Kiran Kumar,2 Patricia C. Babbitt,3,4 and Jacquelyn S. Fetrow1,2* 1Department of Molecular Genetics and Genomics, Wake Forest University, Winston-Salem, North Carolina 27106 2Departments of Computer Science and Physics, Wake Forest University, Winston-Salem, North Carolina 27106 3Department of Bioengineering and Therapeutic Sciences, Institute for Quantitative Biosciences University of California San Francisco, San Francisco, California 94158 4Department of Pharmaceutical Chemistry, Institute for Quantitative Biosciences University of California San Francisco, San Francisco, California 94158 Received 10 April 2015; Accepted 10 June 2015 DOI: 10.1002/pro.2724 Published online 12 June 2015 proteinscience.org Abstract: The development of accurate protein function annotation methods has emerged as a major unsolved biological problem. Protein similarity networks, one approach to function annota- tion via annotation transfer, group proteins into similarity-based clusters. An underlying assump- tion is that the edge metric used to identify such clusters correlates with functional information. In this contribution, this assumption is evaluated by observing topologies in similarity networks using three different edge metrics: sequence (BLAST), structure (TM-Align), and active site similarity (active site profiling, implemented in DASP). Network topologies for four well-studied protein superfamilies (enolase, peroxiredoxin (Prx), glutathione transferase (GST), and crotonase) were compared with curated functional hierarchies and structure. As expected, network topology differs, depending on edge metric; comparison of topologies provides valuable information on structure/ function relationships. Subnetworks based on active site similarity correlate with known functional hierarchies at a single edge threshold more often than sequence- or structure-based networks. -

Functional Assignments in the Enolase Superfamily: Investigations of Two Divergent Groups of D-Galacturonate Dehydratases and Galactarate Dehydratase-Iii
FUNCTIONAL ASSIGNMENTS IN THE ENOLASE SUPERFAMILY: INVESTIGATIONS OF TWO DIVERGENT GROUPS OF D-GALACTURONATE DEHYDRATASES AND GALACTARATE DEHYDRATASE-III BY FIONA PATRICIA GRONINGER-POE i DISSERTATION Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biochemistry in the Graduate College of the University of Illinois at Urbana-Champaign, 2014 Urbana, Illinois Doctoral Committee: Professor John A. Gerlt, Chair Professor John E. Cronan, Jr. Associate Professor Rutilo Fratti Associate Professor Raven H. Huang ABSTRACT More than a decade after the genomic age, full genome sequencing is cost-effective and fast, allowing for the deposit of an ever increasing number of DNA sequences. New fields have arisen from this availability of genomic information, and the way we think about biochemistry and enzymology has been transformed. Unfortunately, there is no robust method for accurately determining the functions of enzymes encoded by these sequences that matches the speed in which genomes are deposited into public databases. Functional assignment of enzymes remains of utmost importance in understanding microbial metabolism and has applications in agriculture by examining bacterial plant pathogen metabolism and additionally in human health by providing metabolic context to the human gut microbiome. To aid in the functional identification of proteins, enzymes can be grouped into superfamilies which share common structural motifs as well as mechanistic features. To this end, the enolase superfamily is an excellent model system for functional assignment because more than half of the members still lack functional identification. Structurally, these enzymes contain substrate specificity residues in the N-terminal capping domain and catalytic residues in the C-terminal barrel domain. -

Supplementary Informations SI2. Supplementary Table 1
Supplementary Informations SI2. Supplementary Table 1. M9, soil, and rhizosphere media composition. LB in Compound Name Exchange Reaction LB in soil LBin M9 rhizosphere H2O EX_cpd00001_e0 -15 -15 -10 O2 EX_cpd00007_e0 -15 -15 -10 Phosphate EX_cpd00009_e0 -15 -15 -10 CO2 EX_cpd00011_e0 -15 -15 0 Ammonia EX_cpd00013_e0 -7.5 -7.5 -10 L-glutamate EX_cpd00023_e0 0 -0.0283302 0 D-glucose EX_cpd00027_e0 -0.61972444 -0.04098397 0 Mn2 EX_cpd00030_e0 -15 -15 -10 Glycine EX_cpd00033_e0 -0.0068175 -0.00693094 0 Zn2 EX_cpd00034_e0 -15 -15 -10 L-alanine EX_cpd00035_e0 -0.02780553 -0.00823049 0 Succinate EX_cpd00036_e0 -0.0056245 -0.12240603 0 L-lysine EX_cpd00039_e0 0 -10 0 L-aspartate EX_cpd00041_e0 0 -0.03205557 0 Sulfate EX_cpd00048_e0 -15 -15 -10 L-arginine EX_cpd00051_e0 -0.0068175 -0.00948672 0 L-serine EX_cpd00054_e0 0 -0.01004986 0 Cu2+ EX_cpd00058_e0 -15 -15 -10 Ca2+ EX_cpd00063_e0 -15 -100 -10 L-ornithine EX_cpd00064_e0 -0.0068175 -0.00831712 0 H+ EX_cpd00067_e0 -15 -15 -10 L-tyrosine EX_cpd00069_e0 -0.0068175 -0.00233919 0 Sucrose EX_cpd00076_e0 0 -0.02049199 0 L-cysteine EX_cpd00084_e0 -0.0068175 0 0 Cl- EX_cpd00099_e0 -15 -15 -10 Glycerol EX_cpd00100_e0 0 0 -10 Biotin EX_cpd00104_e0 -15 -15 0 D-ribose EX_cpd00105_e0 -0.01862144 0 0 L-leucine EX_cpd00107_e0 -0.03596182 -0.00303228 0 D-galactose EX_cpd00108_e0 -0.25290619 -0.18317325 0 L-histidine EX_cpd00119_e0 -0.0068175 -0.00506825 0 L-proline EX_cpd00129_e0 -0.01102953 0 0 L-malate EX_cpd00130_e0 -0.03649016 -0.79413596 0 D-mannose EX_cpd00138_e0 -0.2540567 -0.05436649 0 Co2 EX_cpd00149_e0 -

SI Appendix Index 1
SI Appendix Index Calculating chemical attributes using EC-BLAST ................................................................................ 2 Chemical attributes in isomerase reactions ............................................................................................ 3 Bond changes …..................................................................................................................................... 3 Reaction centres …................................................................................................................................. 5 Substrates and products …..................................................................................................................... 6 Comparative analysis …........................................................................................................................ 7 Racemases and epimerases (EC 5.1) ….................................................................................................. 7 Intramolecular oxidoreductases (EC 5.3) …........................................................................................... 8 Intramolecular transferases (EC 5.4) ….................................................................................................. 9 Supporting references …....................................................................................................................... 10 Fig. S1. Overview …............................................................................................................................ -

Functional and Physiological Discovery in the Mannonate Dehydratase Subgroup of the Enolase Superfamily
FUNCTIONAL AND PHYSIOLOGICAL DISCOVERY IN THE MANNONATE DEHYDRATASE SUBGROUP OF THE ENOLASE SUPERFAMILY BY DANIEL JOSEPH WICHELECKI DISSERTATION Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biochemistry in the Graduate College of the University of Illinois at Urbana-Champaign, 2014 Urbana, Illinois Doctoral Committee: Professor John Gerlt, Chair Professor John Cronan Professor Scott Silverman Professor Wilfred van der Donk ABSTRACT In the current post-genomic world, the exponential amassing of protein sequences is overwhelming the scientific community’s ability to experimentally assign each protein’s function. The use of automated, homology-based annotations has allowed a reprieve from this efflux of data, but has led to widespread misannotation and nonannotation in protein sequence databases. This dissertation details the functional and physiological characterization of the mannonate dehydratase subgroup (ManD) of the enolase superfamily (ENS). The outcome affirms the dangers of homology-based annotations while discovering novel metabolic pathways. Furthermore, the experimental verification of these pathways ( in vitro and in vivo ) has provided a platform to test the general strategies for improved functional and metabolic characterization being developed by the Enzyme Function Initiative (EFI). Prior to this study, one member of the ManD subgroup had been characterized and was shown to dehydrate D-mannonate to 2-keto-3-deoxy-D-gluconate. Forty-two additional members of the ManD, selected from across the sequence space of the subgroup, were screened for activity and kinetic constants were determined. The members of the once isofunctional subgroup were found to differ in both catalytic efficiency and substrate specificity: 1) high 3 4 -1 -1 efficiency (k cat /K M = 10 to 10 M s ) dehydration of D-mannonate, 2) low efficiency (k cat /K M = 10 1 to 10 2 M-1s-1) dehydration of D-mannonate and/or D-gluconate, and 3) no-activity with either D-mannonate or D-gluconate (or any other acid sugar tested). -

Supporting Information for Proteomics DOI 10.1002/Pmic.200600987
Supporting Information for Proteomics DOI 10.1002/pmic.200600987 Lars Whlbrand, Birte Kallerhoff, Daniela Lange, Peter Hufnagel, Jrgen Thiermann, Richard Reinhardt and Ralf Rabus Functional proteomic view of metabolic regulation in “Aromatoleum aromaticum” strain EbN1 ª 2007 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim www.proteomics-journal.com Content 1. General proteome features Page Experimental outline of the presented study with “Aromatoleum aromaticum” 1-2 strain EbN1 (Fig. 1.1.) Spot pattern comparison of differently visualized 2DE gels (Fig. 1.2.) 1-3 Principle component analysis of the proteins for each analyzed 2D DIGE gel 1-4 of all main physiological groups (Fig. 1.3.) Fold changes in protein abundance of all identified proteins (Fig 1-5 Number of regulated as well as not regulated protein spots on the 2DE gels 1-6 and their share of total protein (Table 1.1.) Mean average ratio of not regulated protein spots of the three main 1-7 physiological groups (Table 1.2.) Mean average ratios of not regulated proteins, identified in all three main 1-8 physiological groups (Table 1.3.) Proteins identified in this study (Table 1.4.) 1-9 2. Cellular functions* 2.1. DNA replication, recombination and repair 2-2 2.1.1. DNA replication 2-2 2.2. Transcription 2-4 2.2.1. DNA-dependent RNA polymerase and sigma factors 2-5 2.2.2. Factors affecting RNA polymerase 2-5 2.2.3. Classes of transcriptional regulators 2-5 2.3. Translation 2-6 2.3.1. tRNA synthesis 2-6 2.3.2. tRNA/rRNA modification 2-7 2.3.3. -

Oup Proeng Gzy011 135..145 ++
Protein Engineering, Design & Selection, 2018, vol. 31 no. 4, pp. 135–145 doi: 10.1093/protein/gzy011 Advance Access Publication Date: 30 May 2018 Original Article Original Article A platform for chemical modification of mandelate racemase: characterization of the C92S/C264S and γ-thialysine 166 variants Downloaded from https://academic.oup.com/peds/article/31/4/135/5025590 by guest on 30 September 2021 Mitesh Nagar1, Himank Kumar1, and Stephen L. Bearne1,2,* 1Department of Biochemistry and Molecular Biology, Dalhousie University, Halifax, NS, Canada B3H 4R2, and 2Department of Chemistry, Dalhousie University, Halifax, NS, Canada B3H 4R2 *To whom correspondence should be addressed. E-mail: [email protected] Edited by Miroslaw Cygler Received 7 February 2018; Revised 28 April 2018; Editorial Decision 30 April 2018; Accepted 3 May 2018 Abstract Mandelate racemase (MR) serves as a paradigm for our understanding of enzyme-catalyzed deprotona- tion of a carbon acid substrate. To facilitate structure–function studies on MR using non-natural amino acid substitutions, we engineered the Cys92Ser/Cys264Ser variant (dmMR) as a platform for introdu- cingCysresiduesatspecific locations for subsequent covalent modification. While the highly reactive thiol of Cys furnishes a site for chemical modification, site-specificity requires that other Cys residues be non-reactive or replaced by a non-reactive amino acid, especially if chemical modification is con- ducted under denaturing conditions. The catalytic efficiency of dmMR is reduced only ~2-fold relative to wild-type MR, making dmMR a viable platform for the site-specificintroductionofCys.Asan example, the inactive Lys166Cys variant of dmMR was treated with ethylenimine under denaturing con- ditions to replace the Brønsted acid-base catalyst Lys 166 with the non-natural amino acid γ-thialysine. -

Impact of Temperature on the Time Required for the Establishment of Primordial Biochemistry, and for the Evolution of Enzymes
Impact of temperature on the time required for the establishment of primordial biochemistry, and for the evolution of enzymes Randy B. Stockbridge, Charles A. Lewis, Jr., Yang Yuan, and Richard Wolfenden1 Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC 27599 Contributed by Richard Wolfenden, September 29, 2010 (sent for review August 1, 2010) All reactions are accelerated by an increase in temperature, but the magnitude of that effect on very slow reactions does not seem to have been fully appreciated. The hydrolysis of polysaccharides, for example, is accelerated 190,000-fold when the temperature is raised from 25 to 100 °C, while the rate of hydrolysis of phosphate monoester dianions increases 10,300,000-fold. Moreover, the slow- est reactions tend to be the most heat-sensitive. These tendencies collapse, by as many as five orders of magnitude, the time that would have been required for early chemical evolution in a warm environment. We propose, further, that if the catalytic effect of a “proto-enzyme”—like that of modern enzymes—were mainly enthalpic, then the resulting rate enhancement would have in- creased automatically as the environment became cooler. Several powerful nonenzymatic catalysts of very slow biological reactions, notably pyridoxal phosphate and the ceric ion, are shown to meet that criterion. Taken together, these findings greatly reduce the time that would have been required for early chemical evolution, countering the view that not enough time has passed for life to Fig. 1. Half-lives (t1∕2) and first order rate constants (k) of some biological have evolved to its present level of complexity. -

Novel Non-Phosphorylative Pathway of Pentose Metabolism from Bacteria
www.nature.com/scientificreports OPEN Novel non-phosphorylative pathway of pentose metabolism from bacteria Received: 22 March 2018 Seiya Watanabe1,2,3, Fumiyasu Fukumori4, Hisashi Nishiwaki1,2, Yasuhiro Sakurai5, Accepted: 30 September 2018 Kunihiko Tajima5 & Yasuo Watanabe1,2 Published: xx xx xxxx Pentoses, including D-xylose, L-arabinose, and D-arabinose, are generally phosphorylated to D-xylulose 5-phosphate in bacteria and fungi. However, in non-phosphorylative pathways analogous to the Entner-Dodorof pathway in bacteria and archaea, such pentoses can be converted to pyruvate and glycolaldehyde (Route I) or α-ketoglutarate (Route II) via a 2-keto-3-deoxypentonate (KDP) intermediate. Putative gene clusters related to these metabolic pathways were identifed on the genome of Herbaspirillum huttiense IAM 15032 using a bioinformatic analysis. The biochemical characterization of C785_RS13685, one of the components encoded to D-arabinonate dehydratase, difered from the known acid-sugar dehydratases. The biochemical characterization of the remaining components and a genetic expression analysis revealed that D- and L-KDP were converted not only to α-ketoglutarate, but also pyruvate and glycolate through the participation of dehydrogenase and hydrolase (Route III). Further analyses revealed that the Route II pathway of D-arabinose metabolism was not evolutionally related to the analogous pathway from archaea. Te breakdown of D-glucose is central for energy and biosynthetic metabolism throughout all domains of life. Te most common glycolytic routes in bacteria are the Embden-Meyerhof-Parnas, the Entner-Doudorof (ED), and the oxidative pentose phosphate pathways. Te distinguishing diference between the two former glyco- lytic pathways lies in the nature of the 6-carbon metabolic intermediates that serve as substrates for aldol cleav- age. -
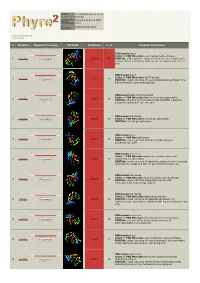
Phyre 2 Results for P29208
Email [email protected] Description P29208 Thu Jan 5 11:45:36 GMT Date 2012 Unique Job 0bd634b8f8620003 ID Detailed template information # Template Alignment Coverage 3D Model Confidence % i.d. Template Information PDB header:lyase Chain: A: PDB Molecule:o-succinylbenzoate synthase; 1 c3gc2A_ Alignment 100.0 86 PDBTitle: 1.85 angstrom crystal structure of o-succinylbenzoate synthase from2 salmonella typhimurium in complex with succinic acid PDB header:lyase Chain: A: PDB Molecule:tlr1174 protein; 2 c2oztA_ 100.0 20 Alignment PDBTitle: crystal structure of o-succinylbenzoate synthase from2 thermosynechococcus elongatus bp-1 PDB header:metal binding protein Chain: A: PDB Molecule:enzyme of enolase superfamily; 3 c3px5A_ 100.0 19 Alignment PDBTitle: structure of efi enolase target en500555, a putative dipeptide2 epimerase: apo structure PDB header:isomerase 4 c1jpmB_ Alignment 100.0 20 Chain: B: PDB Molecule:l-ala-d/l-glu epimerase; PDBTitle: l-ala-d/l-glu epimerase PDB header:lyase Chain: A: PDB Molecule:menc; 5 c2pgeA_ 100.0 23 Alignment PDBTitle: crystal structure of menc from desulfotalea psychrophila2 lsv54 PDB header:isomerase Chain: E: PDB Molecule:mandelate racemase/muconate 6 c3q45E_ Alignment 100.0 19 lactonizing enzyme family; PDBTitle: crystal structure of dipeptide epimerase from cytophaga hutchinsonii2 complexed with mg and dipeptide d-ala-l-val PDB header:isomerase Chain: A: PDB Molecule:chloromuconate cycloisomerase; 7 c1nu5A_ 100.0 16 Alignment PDBTitle: crystal structure of pseudomonas sp. p51 chloromuconate lactonizing2 -

The Enolase Superfamily: a General Strategy for Enzyme-Catalyzed Abstraction of the R-Protons of Carboxylic Acids†
+ + Biochemistry 1996, 35, 16489-16501 16489 The Enolase Superfamily: A General Strategy for Enzyme-Catalyzed Abstraction of the R-Protons of Carboxylic Acids† ,‡ §,| ⊥,# Patricia C. Babbitt,* Miriam S. Hasson, Joseph E. Wedekind, David R. J. Palmer,r William C. Barrett,r ⊥ ⊥ § ‡ , George H. Reed, Ivan Rayment, Dagmar Ringe, George L. Kenyon, and John A. Gerlt* r Department of Pharmaceutical Chemistry, UniVersity of California, San Francisco, California 94143-0446, Departments of Biochemistry and Chemistry and Rosenstiel Center for Basic Biomedical Research, Brandeis UniVersity, Waltham, Massachusetts 02154-9110, The Institute for Enzyme Research and Department of Biochemistry, UniVersity of Wisconsin, Madison, Wisconsin 53706, and Department of Biochemistry, UniVersity of Illinois, Urbana, Illinois 61801 ReceiVed July 5, 1996X ABSTRACT: We have discovered a superfamily of enzymes related by their ability to catalyze the abstraction of the R-proton of a carboxylic acid to form an enolic intermediate. Although each reaction catalyzed by these enzymes is initiated by this common step, their overall reactions (including racemization, â-elimination of water, â-elimination of ammonia, and cycloisomerization) as well as the stereochemical consequences (syn Vs anti) of the â-elimination reactions are diverse. Analysis of sequence and structural similarities among these proteins suggests that all of their chemical reactions are mediated by a common active site architecture modified through evolution to allow the enolic intermediates to partition to different products in their respective active sites Via different overall mechanisms. All of these enzymes retain the ability to catalyze the thermodynamically difficult step of proton abstraction. These homologous proteins, designated the “enolase superfamily”, include enolase as well as more metabolically specialized enzymes: mandelate racemase, galactonate dehydratase, glucarate dehydratase, muconate-lactonizing enzymes, N-acylamino acid racemase, â-methylaspartate ammonia-lyase, and o-succinylbenzoate synthase.