Iraspa: GPU-Accelerated Visualization Software for Materials Scientists
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
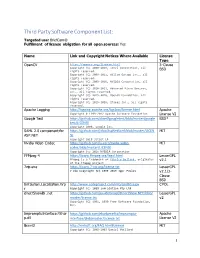
Third Party Software Component List: Targeted Use: Briefcam® Fulfillment of License Obligation for All Open Sources: Yes
Third Party Software Component List: Targeted use: BriefCam® Fulfillment of license obligation for all open sources: Yes Name Link and Copyright Notices Where Available License Type OpenCV https://opencv.org/license.html 3-Clause Copyright (C) 2000-2019, Intel Corporation, all BSD rights reserved. Copyright (C) 2009-2011, Willow Garage Inc., all rights reserved. Copyright (C) 2009-2016, NVIDIA Corporation, all rights reserved. Copyright (C) 2010-2013, Advanced Micro Devices, Inc., all rights reserved. Copyright (C) 2015-2016, OpenCV Foundation, all rights reserved. Copyright (C) 2015-2016, Itseez Inc., all rights reserved. Apache Logging http://logging.apache.org/log4cxx/license.html Apache Copyright © 1999-2012 Apache Software Foundation License V2 Google Test https://github.com/abseil/googletest/blob/master/google BSD* test/LICENSE Copyright 2008, Google Inc. SAML 2.0 component for https://github.com/jitbit/AspNetSaml/blob/master/LICEN MIT ASP.NET SE Copyright 2018 Jitbit LP Nvidia Video Codec https://github.com/lu-zero/nvidia-video- MIT codec/blob/master/LICENSE Copyright (c) 2016 NVIDIA Corporation FFMpeg 4 https://www.ffmpeg.org/legal.html LesserGPL FFmpeg is a trademark of Fabrice Bellard, originator v2.1 of the FFmpeg project 7zip.exe https://www.7-zip.org/license.txt LesserGPL 7-Zip Copyright (C) 1999-2019 Igor Pavlov v2.1/3- Clause BSD Infralution.Localization.Wp http://www.codeproject.com/info/cpol10.aspx CPOL f Copyright (C) 2018 Infralution Pty Ltd directShowlib .net https://github.com/pauldotknopf/DirectShow.NET/blob/ LesserGPL -

A Web-Based 3D Molecular Structure Editor and Visualizer Platform
Mohebifar and Sajadi J Cheminform (2015) 7:56 DOI 10.1186/s13321-015-0101-7 SOFTWARE Open Access Chemozart: a web‑based 3D molecular structure editor and visualizer platform Mohamad Mohebifar* and Fatemehsadat Sajadi Abstract Background: Chemozart is a 3D Molecule editor and visualizer built on top of native web components. It offers an easy to access service, user-friendly graphical interface and modular design. It is a client centric web application which communicates with the server via a representational state transfer style web service. Both client-side and server-side application are written in JavaScript. A combination of JavaScript and HTML is used to draw three-dimen- sional structures of molecules. Results: With the help of WebGL, three-dimensional visualization tool is provided. Using CSS3 and HTML5, a user- friendly interface is composed. More than 30 packages are used to compose this application which adds enough flex- ibility to it to be extended. Molecule structures can be drawn on all types of platforms and is compatible with mobile devices. No installation is required in order to use this application and it can be accessed through the internet. This application can be extended on both server-side and client-side by implementing modules in JavaScript. Molecular compounds are drawn on the HTML5 Canvas element using WebGL context. Conclusions: Chemozart is a chemical platform which is powerful, flexible, and easy to access. It provides an online web-based tool used for chemical visualization along with result oriented optimization for cloud based API (applica- tion programming interface). JavaScript libraries which allow creation of web pages containing interactive three- dimensional molecular structures has also been made available. -

Water and Salt at the Lipid-Solvent Interface
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School April 2019 Water and Salt at the Lipid-Solvent Interface James M. Kruczek University of South Florida, [email protected] Follow this and additional works at: https://scholarcommons.usf.edu/etd Part of the Physics Commons Scholar Commons Citation Kruczek, James M., "Water and Salt at the Lipid-Solvent Interface" (2019). Graduate Theses and Dissertations. https://scholarcommons.usf.edu/etd/8380 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Water and Salt at the Lipid-Solvent Interface by James M. Kruczek A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Applied Physics Department of Physics College of Arts and Sciences University of South Florida Major Professor: Sagar A. Pandit, Ph.D. Ullah, Ghanim, Ph.D. Robert S. Hoy, Ph.D. Jianjun Pan, Ph.D. Yicheng Tu, Ph.D. Date of Approval: March 26, 2019 Keywords: Lipid Bilayer, Ionic Solvents, Ether Lipids, Molecular Simulations Copyright ⃝c 2018, James M. Kruczek Dedication To my wife Nicole, without whom none of this would be possible. To my father Michael, who labored for his family till his last days. To my family, for all of their support. Acknowledgments The work presented in this document would not be possible without the assistance of many academic professionals. In particular, I would like to acknowledge my major advisor Dr. -

Tracking of Bacterial Metabolism with Azidated Precursors and Click- Chemistry”
Tracking of bacterial metabolism with azidated precursors and click-chemistry Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften vorgelegt beim Fachbereich für Biowissenschaften (15) der Johann Wolfang Goethe Universität in Frankfurt am Main von Alexander J. Pérez aus Nürnberg Frankfurt am Main 2015 Dekanin: Prof. Dr. Meike Piepenbring Gutachter: Prof. Dr. Helge B. Bode Zweitgutachter: Prof. Dr. Joachim W. Engels Datum der Disputation: 2 Danksagung Ich danke meinen Eltern für die stete und vielseitige Unterstützung, deren Umfang ich sehr zu schätzen weiß. Herrn Professor Dr. Helge B. Bode gilt mein besonderer Dank für die Übernahme als Doktorand und für die Gelegenheit meinen Horizont in diesen mich stets faszinierenden Themenbereich in dieser Tiefe erweitern zu lassen. Seine persönliche und fachliche Unterstützung bei der Projektwahl und der entsprechenden Umsetzung ist in dieser Form eine Seltenheit und ich bin mir dieser Tatsache voll bewusst. Gerade die zusätzlich erworbenen Kenntnisse im Bereich der Biologie, sowie der Wert interdisziplinärer Zusammenarbeit ist mir durch zahlreiche freundliche und wertvolle Mitglieder der Arbeitsgruppe bewusst geworden und viele zündenden Ideen wären ohne sie womöglich nie aufgekommen. Einen besonderen Dank möchte ich in diesem Kontext Wolfram Lorenzen und Sebastian Fuchs, die gerade in der Anfangszeit eine große Hilfe waren, ausdrücken. Dies gilt ebenso für die „N100-Crew“ und sämtliche Freunde, die in dieser Zeit zu mir standen und diesen Lebensabschnitt unvergesslich gemacht haben. -
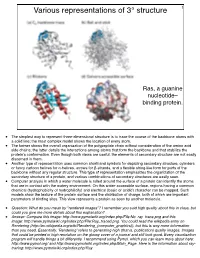
Various Representations of 3° Structure 1
Various representations of 3° structure 1 Ras, a guanine nucleotide– binding protein. • The simplest way to represent three-dimensional structure is to trace the course of the backbone atoms with a solid line; the most complex model shows the location of every atom. • The former shows the overall organization of the polypeptide chain without consideration of the amino acid side chains; the latter details the interactions among atoms that form the backbone and that stabilize the protein’s conformation. Even though both views are useful, the elements of secondary structure are not easily discerned in them. • Another type of representation uses common shorthand symbols for depicting secondary structure, cylinders or fancy cartoon helices for α-helices, arrows for β-strands, and a flexible string-like form for parts of the backbone without any regular structure. This type of representation emphasizes the organization of the secondary structure of a protein, and various combinations of secondary structures are easily seen. • Computer analysis in which a water molecule is rolled around the surface of a protein can identify the atoms that are in contact with the watery environment. On this water-accessible surface, regions having a common chemical (hydrophobicity or hydrophilicity) and electrical (basic or acidic) character can be mapped. Such models show the texture of the protein surface and the distribution of charge, both of which are important parameters of binding sites. This view represents a protein as seen by another molecule. • Question: What do you mean by "rendered images"? I remember you said high quality about this in class, but could you give me more details about this explanation? • Answer: Compare this image: http://www.pymolwiki.org/index.php/File:No_ray_trace.png and this image: http://www.pymolwiki.org/index.php/File:Ray_traced.png. -

The Deep Learning Solutions on Lossless Compression Methods for Alleviating Data Load on Iot Nodes in Smart Cities
sensors Article The Deep Learning Solutions on Lossless Compression Methods for Alleviating Data Load on IoT Nodes in Smart Cities Ammar Nasif *, Zulaiha Ali Othman and Nor Samsiah Sani Center for Artificial Intelligence Technology (CAIT), Faculty of Information Science & Technology, University Kebangsaan Malaysia, Bangi 43600, Malaysia; [email protected] (Z.A.O.); [email protected] (N.S.S.) * Correspondence: [email protected] Abstract: Networking is crucial for smart city projects nowadays, as it offers an environment where people and things are connected. This paper presents a chronology of factors on the development of smart cities, including IoT technologies as network infrastructure. Increasing IoT nodes leads to increasing data flow, which is a potential source of failure for IoT networks. The biggest challenge of IoT networks is that the IoT may have insufficient memory to handle all transaction data within the IoT network. We aim in this paper to propose a potential compression method for reducing IoT network data traffic. Therefore, we investigate various lossless compression algorithms, such as entropy or dictionary-based algorithms, and general compression methods to determine which algorithm or method adheres to the IoT specifications. Furthermore, this study conducts compression experiments using entropy (Huffman, Adaptive Huffman) and Dictionary (LZ77, LZ78) as well as five different types of datasets of the IoT data traffic. Though the above algorithms can alleviate the IoT data traffic, adaptive Huffman gave the best compression algorithm. Therefore, in this paper, Citation: Nasif, A.; Othman, Z.A.; we aim to propose a conceptual compression method for IoT data traffic by improving an adaptive Sani, N.S. -

Visualizing Protein Structures-Tools and Trends
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 12 January 2020 Visualizing protein structures - tools and trends 1,2 3 1,2 X. Martinez , M. Chavent , M. Baaden 1) CNRS, Université de Paris, UPR 9080, Laboratoire de Biochimie Théorique, 13 rue Pierre et Marie Curie, F-75005, Paris, France 2) Institut de Biologie Physico-Chimique-Fondation Edmond de Rotschild, PSL Research University, Paris, France 3) Institut de Pharmacologie et de Biologie Structurale IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France Abstract Molecular visualisation is fundamental in the current scientific literature, textbooks and dissemination materials, forming an essential support for presenting results, reasoning on and formulating hypotheses related to molecular structure. Visual exploration has become easily accessible on a broad variety of platforms thanks to advanced software tools that render a great service to the scientific community. These tools are often developed across disciplines bridging computer science, biology and chemistry. Here we first describe a few Swiss Army knives geared towards protein visualisation for everyday use with an existing large user base, then focus on more specialised tools for peculiar needs that are not yet as broadly known. Our selection is by no means exhaustive, but reflects a diverse snapshot of scenarios that we consider informative for the reader. We end with an account of future trends and perspectives. Keywords Molecular Graphics, Protein visualization, Software tools, Virtual reality Introduction Many parts of science rely on a visualization-driven cycle of experimentation, reasoning, conjecture and validation, even more so in relation with structural biology and biophysics. Molecular visualization (1) in particular is now broadly used in many contexts, with the purpose of illustration in the scientific literature or the aim to gain insight about primary research data. -

Forcepoint DLP Supported File Formats and Size Limits
Forcepoint DLP Supported File Formats and Size Limits Supported File Formats and Size Limits | Forcepoint DLP | v8.8.1 This article provides a list of the file formats that can be analyzed by Forcepoint DLP, file formats from which content and meta data can be extracted, and the file size limits for network, endpoint, and discovery functions. See: ● Supported File Formats ● File Size Limits © 2021 Forcepoint LLC Supported File Formats Supported File Formats and Size Limits | Forcepoint DLP | v8.8.1 The following tables lists the file formats supported by Forcepoint DLP. File formats are in alphabetical order by format group. ● Archive For mats, page 3 ● Backup Formats, page 7 ● Business Intelligence (BI) and Analysis Formats, page 8 ● Computer-Aided Design Formats, page 9 ● Cryptography Formats, page 12 ● Database Formats, page 14 ● Desktop publishing formats, page 16 ● eBook/Audio book formats, page 17 ● Executable formats, page 18 ● Font formats, page 20 ● Graphics formats - general, page 21 ● Graphics formats - vector graphics, page 26 ● Library formats, page 29 ● Log formats, page 30 ● Mail formats, page 31 ● Multimedia formats, page 32 ● Object formats, page 37 ● Presentation formats, page 38 ● Project management formats, page 40 ● Spreadsheet formats, page 41 ● Text and markup formats, page 43 ● Word processing formats, page 45 ● Miscellaneous formats, page 53 Supported file formats are added and updated frequently. Key to support tables Symbol Description Y The format is supported N The format is not supported P Partial metadata -

Engine for Molecule Visualization in a Web Browser
MASARYKOVA UNIVERZITA FAKULTA}w¡¢£¤¥¦§¨ INFORMATIKY !"#$%&'()+,-./012345<yA| Engine for Molecule Visualization in a Web Browser MASTER’S THESIS Jaromír Svoboda Brno, spring 2014 Declaration Hereby I declare, that this paper is my original authorial work, which I have worked out by my own. All sources, references and literature used or excerpted during elaboration of this work are properly cited and listed in complete reference to the due source. Jaromír Svoboda Advisor: RNDr. David Sehnal ii Acknowledgement I would like to thank my supervisor RNDr. David Sehnal for patient guidance and informed advice throughout writing this thesis. iii Abstract The main focus of this master’s thesis is the design and implementa- tion of lightweight molecular visualization engine (called LiveMol) in form of a JavaScript library. The engine utilizes widely adopted WebGL API to display GPU-accelerated graphics in web browsers. Due to the size of complex protein molecules, the primary goal is high performance. Furthermore, the design of LiveMol enables users to extend the core functionality by simply defining custom coloring schemes of molecule models or implement completely new visual- ization modes. iv Keywords molecular visualization, protein, secondary structure, WebGL, LiveMol v Contents I INTRODUCTION 1 1 Introduction ............................2 II CURRENT DEVELOPMENTS 3 2 Current Molecular Visualization Software ..........4 2.1 Desktop Applications ....................4 2.1.1 Visual Molecular Dynamic . .4 2.1.2 PyMOL . .4 2.1.3 RasMol/OpenRasMol . .5 2.1.4 BALL/BALLView . .5 2.1.5 Gabedit . .5 2.1.6 QuteMol . .5 2.1.7 Avogadro . .6 2.2 JavaScript-based Web Applications ............6 2.2.1 Jmol/JSmol . -

Illustrative Molecular Visualization with Continuous Abstraction Matthew Van Der Zwan, Wouter Lueks, Henk Bekker, Tobias Isenberg
Illustrative Molecular Visualization with Continuous Abstraction Matthew van der Zwan, Wouter Lueks, Henk Bekker, Tobias Isenberg To cite this version: Matthew van der Zwan, Wouter Lueks, Henk Bekker, Tobias Isenberg. Illustrative Molecular Visu- alization with Continuous Abstraction. Computer Graphics Forum, Wiley, 2011, 30 (3), pp.683-690. 10.1111/j.1467-8659.2011.01917.x. hal-00781508 HAL Id: hal-00781508 https://hal.inria.fr/hal-00781508 Submitted on 27 Jan 2013 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Public Domain Eurographics / IEEE Symposium on Visualization 2011 (EuroVis 2011) Volume 30 (2011), Number 3 H. Hauser, H. Pfister, and J. J. van Wijk (Guest Editors) Illustrative Molecular Visualization with Continuous Abstraction Matthew van der Zwan,1 Wouter Lueks,1 Henk Bekker,1 and Tobias Isenberg1,2 1Johann Bernoulli Institute of Mathematics and Computer Science, University of Groningen, The Netherlands 2DIGITEO in collaboration with VENISE–LIMSI–CNRS and AVIZ–INRIA, Saclay, France Abstract Molecular systems may be visualized with various degrees of structural abstraction, support of spatial perception, and ‘illustrativeness.’In this work we propose and realize methods to create seamless transformations that allow us to affect and change each of these three parameters individually. -

In-Core Compression: How to Shrink Your Database Size in Several Times
In-core compression: how to shrink your database size in several times Aleksander Alekseev Anastasia Lubennikova www.postgrespro.ru Agenda ● What does Postgres store? • A couple of words about storage internals ● Check list for your schema • A set of tricks to optimize database size ● In-core block level compression • Out-of-box feature of Postgres Pro EE ● ZSON • Extension for transparent JSONB compression What this talk doesn’t cover ● MVCC bloat • Tune autovacuum properly • Drop unused indexes • Use pg_repack • Try pg_squeeze ● Catalog bloat • Create less temporary tables ● WAL-log size • Enable wal_compression ● FS level compression • ZFS, btrfs, etc Data layout Empty tables are not that empty ● Imagine we have no data create table tbl(); insert into tbl select from generate_series(0,1e07); select pg_size_pretty(pg_relation_size('tbl')); pg_size_pretty --------------- ??? Empty tables are not that empty ● Imagine we have no data create table tbl(); insert into tbl select from generate_series(0,1e07); select pg_size_pretty(pg_relation_size('tbl')); pg_size_pretty --------------- 268 MB Meta information db=# select * from heap_page_items(get_raw_page('tbl',0)); -[ RECORD 1 ]------------------- lp | 1 lp_off | 8160 lp_flags | 1 lp_len | 32 t_xmin | 720 t_xmax | 0 t_field3 | 0 t_ctid | (0,1) t_infomask2 | 2 t_infomask | 2048 t_hoff | 24 t_bits | t_oid | t_data | Order matters ● Attributes must be aligned inside the row create table bad (i1 int, b1 bigint, i1 int); create table good (i1 int, i1 int, b1 bigint); Safe up to 20% of space. -

Open Source Molecular Modeling
Accepted Manuscript Title: Open Source Molecular Modeling Author: Somayeh Pirhadi Jocelyn Sunseri David Ryan Koes PII: S1093-3263(16)30118-8 DOI: http://dx.doi.org/doi:10.1016/j.jmgm.2016.07.008 Reference: JMG 6730 To appear in: Journal of Molecular Graphics and Modelling Received date: 4-5-2016 Accepted date: 25-7-2016 Please cite this article as: Somayeh Pirhadi, Jocelyn Sunseri, David Ryan Koes, Open Source Molecular Modeling, <![CDATA[Journal of Molecular Graphics and Modelling]]> (2016), http://dx.doi.org/10.1016/j.jmgm.2016.07.008 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. Open Source Molecular Modeling Somayeh Pirhadia, Jocelyn Sunseria, David Ryan Koesa,∗ aDepartment of Computational and Systems Biology, University of Pittsburgh Abstract The success of molecular modeling and computational chemistry efforts are, by definition, de- pendent on quality software applications. Open source software development provides many advantages to users of modeling applications, not the least of which is that the software is free and completely extendable. In this review we categorize, enumerate, and describe available open source software packages for molecular modeling and computational chemistry. 1. Introduction What is Open Source? Free and open source software (FOSS) is software that is both considered \free software," as defined by the Free Software Foundation (http://fsf.org) and \open source," as defined by the Open Source Initiative (http://opensource.org).