SGM Meeting Abstracts: University of Warwick, 3-6 April 2006
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Universidade Federal Do Rio Grande Do Sul Centro De Biotecnologia Programa De Pós-Graduação Em Biologia Celular E Molecular
UNIVERSIDADE FEDERAL DO RIO GRANDE DO SUL CENTRO DE BIOTECNOLOGIA PROGRAMA DE PÓS-GRADUAÇÃO EM BIOLOGIA CELULAR E MOLECULAR Caracterização Molecular do Microbioma Hospitalar por Sequenciamento de Alto Desempenho Tese de Doutorado Pabulo Henrique Rampelotto Porto Alegre 2019 UNIVERSIDADE FEDERAL DO RIO GRANDE DO SUL CENTRO DE BIOTECNOLOGIA PROGRAMA DE PÓS-GRADUAÇÃO EM BIOLOGIA CELULAR E MOLECULAR Caracterização Molecular do Microbioma Hospitalar por Sequenciamento de Alto Desempenho Tese submetida ao Programa de Pós-Graduação em Biologia Celular e Molecular da UFRGS, como requisito parcial para a obtenção do grau de Doutor em Ciências Pabulo Henrique Rampelotto Orientador: Dr. Rogério Margis Porto Alegre, Abril de 2019 Instituições e fontes financiadoras: Instituições: Laboratório de Genomas e Populações de Plantas (LGPP), Departamento de Biofísica, UFRGS – Porto Alegre/RS, Brasil. Neoprospecta Microbiome Technologies SA – Florianópolis/SC, Brasil. Hospital Universitário Polydoro Ernani de São Thiago, Universidade Federal de Santa Catarina (UFSC) – Florianópolis/SC, Brasil. Fontes financiadoras: Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brasil. Agradecimentos Aos meus familiares, pelo suporte incondicional em todos os momentos de minha vida. Ao meu orientador Prof. Rogério Margis, pela oportunidade e confiança. Aos colegas de laboratório, pelo apoio e amizade. Ao Programa de Pós-Graduação em Biologia Celular e Molecular, por todo o suporte. Aos inúmeros autores e co-autores que participaram dos meus diversos projetos editoriais, pelas brilhantes discussões em temas tão fascinantes. Enfim, a todos que, de alguma forma, contribuíram para a realização deste trabalho. “A tarefa não é tanto ver aquilo que ninguém viu, mas pensar o que ninguém ainda pensou sobre aquilo que todo mundo vê” (Arthur Schopenhauer) SUMÁRIO LISTA DE ABREVIATURAS .......................................................................................... -

A Historical Analysis of Herpes Simplex Virus Promoter Activation in Vivo Reveals Distinct Populations of Latently Infected Neurones
Journal of General Virology (2008), 89, 2965–2974 DOI 10.1099/vir.0.2008/005066-0 A historical analysis of herpes simplex virus promoter activation in vivo reveals distinct populations of latently infected neurones Joa˜o T. Proenc¸a,1 Heather M. Coleman,1 Viv Connor,1 Douglas J. Winton2 and Stacey Efstathiou1 Correspondence 1Division of Virology, Department of Pathology, University of Cambridge, Tennis Court Road, S. Efstathiou Cambridge CB2 1QP, UK [email protected] 2Cancer Research UK Cambridge Research Institute, Li Ka Shing Centre, Robinson Way, Cambridge CB2 0RE, UK Herpes simplex virus type 1 (HSV-1) has the capacity to establish a life-long latent infection in sensory neurones and also to periodically reactivate from these cells. Since mutant viruses defective for immediate-early (IE) expression retain the capacity for latency establishment it is widely assumed that latency is the consequence of a block in IE gene expression. However, it is not clear whether viral gene expression can precede latency establishment following wild-type virus infection. In order to address this question we have utilized a reporter mouse model system to facilitate a historical analysis of viral promoter activation in vivo. This system utilizes recombinant viruses expressing Cre recombinase under the control of different viral promoters and the Cre reporter mouse strain ROSA26R. In this model, viral promoter-driven Cre recombinase mediates a permanent genetic change, resulting in reporter gene activation and permanent marking of latently infected cells. The analyses of HSV-1 recombinants containing human cytomegalovirus major immediate-early, ICP0, gC or latency-associated transcript Received 20 June 2008 promoters linked to Cre recombinase in this system have revealed the existence of a population of Accepted 4 September 2008 neurones that have experienced IE promoter activation prior to the establishment of latency. -

Annual Conference Abstracts
ANNUAL CONFERENCE 14-17 April 2014 Arena and Convention Centre, Liverpool ABSTRACTS SGM ANNUAL CONFERENCE APRIL 2014 ABSTRACTS (LI00Mo1210) – SGM Prize Medal Lecture (LI00Tu1210) – Marjory Stephenson Climate Change, Oceans, and Infectious Disease Prize Lecture Dr. Rita R. Colwell Understanding the basis of antibiotic resistance University of Maryland, College Park, MD, USA as a platform for early drug discovery During the mid-1980s, satellite sensors were developed to monitor Laura JV Piddock land and oceans for purposes of understanding climate, weather, School of Immunity & Infection and Institute of Microbiology and and vegetation distribution and seasonal variations. Subsequently Infection, University of Birmingham, UK inter-relationships of the environment and infectious diseases Antibiotic resistant bacteria are one of the greatest threats to human were investigated, both qualitatively and quantitatively, with health. Resistance can be mediated by numerous mechanisms documentation of the seasonality of diseases, notably malaria including mutations conferring changes to the genes encoding the and cholera by epidemiologists. The new research revealed a very target proteins as well as RND efflux pumps, which confer innate close interaction of the environment and many other infectious multi-drug resistance (MDR) to bacteria. The production of efflux diseases. With satellite sensors, these relationships were pumps can be increased, usually due to mutations in regulatory quantified and comparatively analyzed. More recent studies of genes, and this confers resistance to antibiotics that are often used epidemic diseases have provided models, both retrospective and to treat infections by Gram negative bacteria. RND MDR efflux prospective, for understanding and predicting disease epidemics, systems not only confer antibiotic resistance, but altered expression notably vector borne diseases. -

The Phagotrophic Origin of Eukaryotes and Phylogenetic Classification Of
International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 DOI: 10.1099/ijs.0.02058-0 The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa Department of Zoology, T. Cavalier-Smith University of Oxford, South Parks Road, Oxford OX1 3PS, UK Tel: j44 1865 281065. Fax: j44 1865 281310. e-mail: tom.cavalier-smith!zoo.ox.ac.uk Eukaryotes and archaebacteria form the clade neomura and are sisters, as shown decisively by genes fragmented only in archaebacteria and by many sequence trees. This sisterhood refutes all theories that eukaryotes originated by merging an archaebacterium and an α-proteobacterium, which also fail to account for numerous features shared specifically by eukaryotes and actinobacteria. I revise the phagotrophy theory of eukaryote origins by arguing that the essentially autogenous origins of most eukaryotic cell properties (phagotrophy, endomembrane system including peroxisomes, cytoskeleton, nucleus, mitosis and sex) partially overlapped and were synergistic with the symbiogenetic origin of mitochondria from an α-proteobacterium. These radical innovations occurred in a derivative of the neomuran common ancestor, which itself had evolved immediately prior to the divergence of eukaryotes and archaebacteria by drastic alterations to its eubacterial ancestor, an actinobacterial posibacterium able to make sterols, by replacing murein peptidoglycan by N-linked glycoproteins and a multitude of other shared neomuran novelties. The conversion of the rigid neomuran wall into a flexible surface coat and the associated origin of phagotrophy were instrumental in the evolution of the endomembrane system, cytoskeleton, nuclear organization and division and sexual life-cycles. Cilia evolved not by symbiogenesis but by autogenous specialization of the cytoskeleton. -
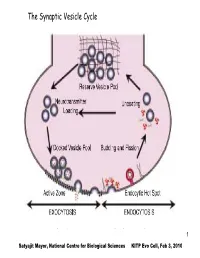
The Synaptic Vesicle Cycle
The Synaptic Vesicle Cycle 1 Satyajit Mayor, National Centre for Biological Sciences KITP Evo Cell, Feb 3, 2010 The Eukaryotic Commandments: T. Cavalier-Smith (i)origin of the endomembrane system (ER, Golgi and lysosomes) and coated- vesicle budding and fusion, including endocytosis and exocytosis; (ii) origin of the cytoskeleton, centrioles, cilia and associated molecular motors; (iii) origin of the nucleus, nuclear pore complex and trans-envelope protein and RNA transport; (iv) origin of linear chromosomes with plural replicons, centromeres and telomeres; (v) origin of novel cell-cycle controls and mitotic segregation; (vi) origin of sex (syngamy, nuclear fusion and meiosis); (vii) origin of peroxisomes; (ix) origin of mitochondria; (viii) novel patterns of rRNAprocessing using small nucleolar-ribonucleoproteins (snoRNPs); (x) origin of spliceosomal introns. 2 EM of Chemical synapse mitochondria Active Zone Figure 5.3, Bear, 2001 3 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. I phase: The ancestors of eukaryotes, the stem neomura, are shared with archaebacteria and evolved during the neomuran revolution, in which N-linked glycoproteins replaced murein peptidoglycan and 18 other suites of characters changed radically through adaptation of an ancestral actinobacterium to thermophily, as discussed in detail by Cavalier- Smith (2002a). 4 T. Cavalier-Smith International Journal of Systematic and Evolutionary Microbiology (2002), 52, 297–354 Fig. 1. The bacterial origins of eukaryotes as a two-stage process. II phase: In the next phase, archaebacteria and eukaryotes diverged dramatically. Archaebacteria retained the wall and therefore their general bacterial cell and genetic organization, but became adapted to even hotter and more acidic environments by substituting prenyl ether lipids for the ancestral acyl esters and making new acid- resistant flagellar shafts (Cavalier-Smith, 2002a). -

Extensive Microbial Diversity Within the Chicken Gut Microbiome Revealed by Metagenomics and Culture
Extensive microbial diversity within the chicken gut microbiome revealed by metagenomics and culture Rachel Gilroy1, Anuradha Ravi1, Maria Getino2, Isabella Pursley2, Daniel L. Horton2, Nabil-Fareed Alikhan1, Dave Baker1, Karim Gharbi3, Neil Hall3,4, Mick Watson5, Evelien M. Adriaenssens1, Ebenezer Foster-Nyarko1, Sheikh Jarju6, Arss Secka7, Martin Antonio6, Aharon Oren8, Roy R. Chaudhuri9, Roberto La Ragione2, Falk Hildebrand1,3 and Mark J. Pallen1,2,4 1 Quadram Institute Bioscience, Norwich, UK 2 School of Veterinary Medicine, University of Surrey, Guildford, UK 3 Earlham Institute, Norwich Research Park, Norwich, UK 4 University of East Anglia, Norwich, UK 5 Roslin Institute, University of Edinburgh, Edinburgh, UK 6 Medical Research Council Unit The Gambia at the London School of Hygiene and Tropical Medicine, Atlantic Boulevard, Banjul, The Gambia 7 West Africa Livestock Innovation Centre, Banjul, The Gambia 8 Department of Plant and Environmental Sciences, The Alexander Silberman Institute of Life Sciences, Edmond J. Safra Campus, Hebrew University of Jerusalem, Jerusalem, Israel 9 Department of Molecular Biology and Biotechnology, University of Sheffield, Sheffield, UK ABSTRACT Background: The chicken is the most abundant food animal in the world. However, despite its importance, the chicken gut microbiome remains largely undefined. Here, we exploit culture-independent and culture-dependent approaches to reveal extensive taxonomic diversity within this complex microbial community. Results: We performed metagenomic sequencing of fifty chicken faecal samples from Submitted 4 December 2020 two breeds and analysed these, alongside all (n = 582) relevant publicly available Accepted 22 January 2021 chicken metagenomes, to cluster over 20 million non-redundant genes and to Published 6 April 2021 construct over 5,500 metagenome-assembled bacterial genomes. -

1985517720.Pdf
JOURNAL OF BACTERIOLOGY, Jan. 2009, p. 347–354 Vol. 191, No. 1 0021-9193/09/$08.00ϩ0 doi:10.1128/JB.01238-08 Copyright © 2009, American Society for Microbiology. All Rights Reserved. Complete Genome Sequence and Comparative Genome Analysis of Enteropathogenic Escherichia coli O127:H6 Strain E2348/69ᰔ† Atsushi Iguchi,1 Nicholas R. Thomson,2 Yoshitoshi Ogura,1,3 David Saunders,2 Tadasuke Ooka,3 Ian R. Henderson,4 David Harris,2 M. Asadulghani,1 Ken Kurokawa,5 Paul Dean,6 Brendan Kenny,6 Michael A. Quail,2 Scott Thurston,2 Gordon Dougan,2 Tetsuya Hayashi,1,3 Julian Parkhill,2 and Gad Frankel7* Division of Bioenvironmental Science, Frontier Science Research Center,1 and Division of Microbiology, Department of Infectious Diseases, Faculty of Medicine,3 University of Miyazaki, Miyazaki, Japan; Pathogen Genomics, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, United Kingdom2; School of Immunity and Infection, University of Downloaded from Birmingham, Birmingham, United Kingdom4; Department of Biological Information, School and Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology, Kanagawa, Japan5; Institute of Cell and Molecular Biosciences, University of Newcastle, Newcastle upon Tyne, United Kingdom6; and Centre for Molecular Microbiology and Infection, Division of Cell and Molecular Biology, Imperial College London, London, United Kingdom7 Received 5 September 2008/Accepted 15 October 2008 Enteropathogenic Escherichia coli (EPEC) was the first pathovar of E. coli to be implicated in human disease; however, no EPEC strain has been fully sequenced until now. Strain E2348/69 (serotype O127:H6 belonging to E. http://jb.asm.org/ coli phylogroup B2) has been used worldwide as a prototype strain to study EPEC biology, genetics, and virulence. -

Holding Hands with Bacteria the Life and Work of Marjory Stephenson
SPRINGER BRIEFS IN MOLECULAR SCIENCE HISTORY OF CHEMISTRY Soňa Štrbáňová Holding Hands with Bacteria The Life and Work of Marjory Stephenson 123 SpringerBriefs in Molecular Science History of Chemistry Series editor Seth C. Rasmussen, Fargo, North Dakota, USA More information about this series at http://www.springer.com/series/10127 Soňa Štrbáňová Holding Hands with Bacteria The Life and Work of Marjory Stephenson 1 3 Soňa Štrbáňová Academy of Sciences of the Czech Republic Prague Czech Republic ISSN 2191-5407 ISSN 2191-5415 (electronic) SpringerBriefs in Molecular Science ISSN 2212-991X SpringerBriefs in History of Chemistry ISBN 978-3-662-49734-0 ISBN 978-3-662-49736-4 (eBook) DOI 10.1007/978-3-662-49736-4 Library of Congress Control Number: 2016934952 © The Author(s) 2016 This work is subject to copyright. All rights are reserved by the Publisher, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilms or in any other physical way, and transmission or information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed. The use of general descriptive names, registered names, trademarks, service marks, etc. in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. The publisher, the authors and the editors are safe to assume that the advice and information in this book are believed to be true and accurate at the date of publication. -

MIAMI UNIVERSITY the Graduate School
MIAMI UNIVERSITY The Graduate School Certificate for Approving the Dissertation We hereby approve the Dissertation of Steven Lindau Distelhorst Candidate for the Degree Doctor of Philosophy ______________________________________ Dr. Mitchell F. Balish, Director ______________________________________ Kelly Z. Abshire, Reader ______________________________________ Natosha L. Finley, Reader ______________________________________ Joseph M. Carlin, Reader ______________________________________ Jack C. Vaughn, Graduate School Representative ABSTRACT UNDERSTANDING VIRULENCE FACTORS OF MYCOPLASMA PENETRANS: ATTACHMENT ORGANELLE ORGANIZATION AND GENE EXPRESSION by Steven Lindau Distelhorst The ability to establish and maintain cell polarity plays an important role in cellular organization for both functional and morphological integrity in eukaryotic and prokaryotic organisms. Like eukaryotes, bacteria, including the genomically reduced species of the Mycoplasma genus, use an array of cytoskeletal proteins to generate and maintain cellular polarity. Some mycoplasmas, such as Mycoplasma penetrans, exhibit a distinct polarized structure, known as the attachment organelle (AO), which is used for attachment to host cells and motility. The M. penetrans AO, like AOs of other mycoplasmas, contains a cytoskeletal structure at the core, but lacks any homologs of identified AO core proteins of other investigated mycoplasmas. To characterize the composition of the M. penetrans AO cytoskeleton we purified the detergent-insoluble core material and examined -

Microbiology Society Activities
MICROBIOLOGY TODAY 48:1 May 2021 Microbiology Today 48:1 May 2021 Life on a Changing Planet Life on a Changing Planet Exploring the relationship between climate change and the microbial world Forest Fire in Dixie National Forest, Utah, USA. santirf/iStock which in turn increase the risk of COVID-19 complications. impact on respiratory health of affected communities. Curr Opin Pulm Wildfire smoke exposure might also increase the likelihood of Med 2019;25:179–187. comorbidities, including cardiovascular diseases, respiratory Wu X, Nethery RC, Sabath MB, Braun D, Dominici F. Air pollution and diseases, diabetes and hypertension, leading to an increased COVID-19 mortality in the United States: strengths and limitations of an ecological regression analysis. Sci Adv 2020;6:eabd4049. COVID-19 risk. Higher daily average PM2.5 concentrations during the wildfire season in western US states were positively Gosling SN, Bryce EK, Dixon PG, Gabriel KMA, Gosling EY et al. associated with increased influenza rates the following A glossary for biometeorology. Int J Biometeorol 2014;58:277–308. winter, indicating strong evidence for delayed effects of wildfire exposure. Research on understanding the underlying mechanisms between wildfire exposure and respiratory Rosa von Borries infection is an area of active investigation. Consultant at World Meteorological Organization, In particular, vulnerable population groups such as socio- Bernhard-Lichtenberg-Str 15, 10407 Berlin, economically disadvantaged populations, individuals with Germany comorbidities and the elderly face higher risks of experiencing [email protected] the combined consequences of COVID-19 and wildfires. @rosavborries In the US, migrant farmworkers who were continuously linkedin.com/in/rosa-von-borries-5b4987197 exposed to wildfire smoke and deemed essential workers during the pandemic faced much higher risks of severe Rosa von Borries holds an MSc in Public Health from London School health consequences from wildfire smoke and COVID-19. -

Mycoplasma Pneumoniae Terminal Organelle
MYCOPLASMA PNEUMONIAE TERMINAL ORGANELLE DEVELOPMENT AND GLIDING MOTILITY by BENJAMIN MICHAEL HASSELBRING (Under the Direction of Duncan Charles Krause) ABSTRACT With a minimal genome containing less than 700 open reading frames and a cell volume < 10% of that of model prokaryotes, Mycoplasma pneumoniae is considered among the smallest and simplest organisms capable of self-replication. And yet, this unique wall-less bacterium exhibits a remarkable level of cellular complexity with a dynamic cytoskeleton and a morphological asymmetry highlighted by a polar, membrane-bound terminal organelle containing an elaborate macromolecular core. The M. pneumoniae terminal organelle functions in distinct, and seemingly disparate cellular processes that include cytadherence, cell division, and presumably gliding motility, as individual cells translocate over surfaces with the cell pole harboring the structure engaged as the leading end. While recent years have witnessed a dramatic increase in the knowledge of protein interactions required for core stability and adhesin trafficking, the mechanism of M. pneumoniae gliding has not been defined nor have interdependencies between the various terminal organelle functions been assessed. The studies presented in the current volume describe the first genetic and molecular investigations into the location, components, architecture, and regulation of the M. pneumoniae gliding machinery. The data indicate that cytadherence and gliding motility are separable properties, and identify a subset of M. pneumoniae proteins contributing directly to the latter process. Characterizations of novel gliding-deficient mutants confirm that the terminal organelle contains the molecular gliding machinery, revealing that with the loss of a single terminal organelle cytoskeletal element, protein P41, terminal organelles detach from the cell body but retain gliding function. -

Quadram Institute Newsletter
Autumn 2019 Welcome to the newsle�er of the Quadram Ins�tute. This issue highlights recent research breakthroughs at the We con�nue to build our team and are pleased to welcome Quadram Ins�tute (QI) that could have posi�ve impacts on Professor Cynthia Whitchurch to the QI. Cynthia is se�ng public health. Working with clinicians our researchers have up a research group inves�ga�ng bacterial lifestyles, and shown how the latest sequencing technologies can aid in how these make them more infec�ous or resistant to diagnos�cs and surveillance. Coupling these techniques an�microbials. Cynthia joins us from the ithree ins�tute at with ‘Big Data’ analy�cal approaches will be vital to the University of Technology Sydney. Her research led to addressing 21st century popula�on health challenges, and the discovery that extracellular DNA is required for biofilm the QI aims to be a leader in this area. development. Big Data in the NHS was the topic of a roundtable We con�nue to welcome visitors to our new building to discussion I a�ended with Patricia Hart at the policy share our vision to understand how food and microbes development think tank Reform. This roundtable was interact to promote health and prevent disease. In June, we sponsored by QI and explored how universi�es and industry had the honour of hos�ng His Excellency Simon Smits, can work with government to realise poten�al applica�ons Ambassador of the Netherlands to the UK. The Ambassador of Big Data. The event was chaired by Baroness Blackwood, visited the Norwich Research Park as part of an Parliamentary Under-Secretary of State, Department of interna�onal trade delega�on to East Anglia to learn about Health and Social Care (DHSC) and included senior the region’s world-leading life sciences research and trade policy-makers, public service prac��oners, academics and opportuni�es.