Central Nervous System (CNS) Transcriptomic Correlates of Human
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Anti-IDH3A Antibody (ARG42205)
Product datasheet [email protected] ARG42205 Package: 100 μl anti-IDH3A antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes IDH3A Tested Reactivity Hu, Ms, Rat Tested Application ICC/IF, IHC-P, WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name IDH3A Antigen Species Human Immunogen Recombinant fusion protein corresponding to aa. 28-366 of Human IDH3A (NP_005521.1). Conjugation Un-conjugated Alternate Names Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial; EC 1.1.1.41; NAD; Isocitric dehydrogenase subunit alpha; + Application Instructions Application table Application Dilution ICC/IF 1:50 - 1:200 IHC-P 1:50 - 1:200 WB 1:500 - 1:2000 Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control Raji Calculated Mw 40 kDa Observed Size ~ 40 kDa Properties Form Liquid Purification Affinity purified. Buffer PBS (pH 7.3), 0.02% Sodium azide and 50% Glycerol. Preservative 0.02% Sodium azide Stabilizer 50% Glycerol Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw www.arigobio.com 1/3 cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Gene Symbol IDH3A Gene Full Name isocitrate dehydrogenase 3 (NAD+) alpha Background Isocitrate dehydrogenases catalyze the oxidative decarboxylation of isocitrate to 2-oxoglutarate. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Reprogramming of Isocitrate Dehydrogenases Expression and Activity by the Androgen
Author Manuscript Published OnlineFirst on May 8, 2019; DOI: 10.1158/1541-7786.MCR-19-0020 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. 1 Reprogramming of isocitrate dehydrogenases expression and activity by the androgen 2 receptor in prostate cancer 3 4 Kevin Gonthier1,2, Raghavendra Tejo Karthik Poluri1,2, Cindy Weidmann1, Maude Tadros1, and 5 Étienne Audet-Walsh1,2,3 6 7 1 Endocrinology - Nephrology Research Axis, Centre de recherche du CHU de Québec - 8 Université Laval, Québec City, Canada 9 2 Department of molecular medicine, Faculty of Medicine, Université Laval, Québec City, 10 Canada 11 3 Centre de recherche sur le cancer de l’Université Laval, Québec City, Canada 12 13 Keywords: steroid; metabolism; glioma; nuclear receptor; castration-resistant prostate cancer, 14 IDH1, oncometabolism 15 16 Abbreviated title: Reprogramming of IDH activity by AR in PCa 17 18 Corresponding Author: Étienne Audet-Walsh, Centre de recherche du CHU de Québec, 2705 19 Boulevard Laurier, room R-4714, Québec City, QC, Canada, G1V 4G2 20 Phone: 1-418-525-4444 ext. 48678 ; e-mail : [email protected] 21 22 Disclosure statement: the authors declare no potential conflicts of interest. 1 Downloaded from mcr.aacrjournals.org on September 24, 2021. © 2019 American Association for Cancer Research. Author Manuscript Published OnlineFirst on May 8, 2019; DOI: 10.1158/1541-7786.MCR-19-0020 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. 23 Abstract 24 Mutations of the isocitrate dehydrogenase genes IDH1 and IDH2, key enzymes involved in 25 citrate metabolism, are important oncogenic events in several cancer types, including in 1-3% of 26 all prostate cancer (PCa) cases. -
Supplementary Data Vigneswaran Et Al Supplementary Data
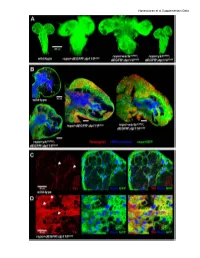
Vigneswaran et al Supplementary Data Vigneswaran et al Supplementary Data Figure S1: Yki is required for EGFR-PI3K-driven glial neoplasia in Drosophila (A) Optical projections of whole brain-nerve cord complexes from 3rd instar larvae approximately 130 hrs old. Dorsal view; anterior up. CD8-GFP (green) labels glial cell bodies. Compared to repo>dEGFRλ;dp110CAAX, warts knockdown (repo>wartsdsRNA; dEGFRλ;dp110CAAX) increased neoplastic brain overgrowth and yki knockdown (repo>ykidsRNA;dEGFRλ;dp110CAAX) decreased neoplastic brain overgrowth. (B) 3 µm optical projections of brain hemispheres, age-matched 3rd instar larvae. Frontal sections; anterior up; midline to left. Repo (red) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies; anti-HRP (blue) counter-stains for neurons and neuropil. (middle) repo>dEGFRλ;dp110CAAX showed increased glial cell numbers (red nuclei) compared to (upper left) wild-type. Compared to repo>dEGFRλ;dp110CAAX, (right) warts knockdown increased neoplastic glial cell numbers (red nuclei), whereas (lower left) yki knockdown reduced neoplastic glial cell numbers (red nuclei). (C, D) Low levels of Yki protein (red) was observed in wild-type central brain glia (white arrows, left panel in C) compared to high levels of cytoplasmic and nuclear Yki protein in dEGFRλ;dp110CAAX neoplastic glia (white arrows, left panel in D); Repo (blue) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies. Vigneswaran et al Supplementary Data Figure S2: YAP/TAZ expression confined to RTK-amplified tuMor cells and Maintained in patient-derived xenografts (A) On the left, immunohistochemical (IHC) staining in representative normal brain parenchyma in the cortex where YAP expression and TAZ expression was limited to vascular cells and was not detectable in normal neuronal and glial cells. -

A Yeast Phenomic Model for the Influence of Warburg Metabolism on Genetic Buffering of Doxorubicin Sean M
Santos and Hartman Cancer & Metabolism (2019) 7:9 https://doi.org/10.1186/s40170-019-0201-3 RESEARCH Open Access A yeast phenomic model for the influence of Warburg metabolism on genetic buffering of doxorubicin Sean M. Santos and John L. Hartman IV* Abstract Background: The influence of the Warburg phenomenon on chemotherapy response is unknown. Saccharomyces cerevisiae mimics the Warburg effect, repressing respiration in the presence of adequate glucose. Yeast phenomic experiments were conducted to assess potential influences of Warburg metabolism on gene-drug interaction underlying the cellular response to doxorubicin. Homologous genes from yeast phenomic and cancer pharmacogenomics data were analyzed to infer evolutionary conservation of gene-drug interaction and predict therapeutic relevance. Methods: Cell proliferation phenotypes (CPPs) of the yeast gene knockout/knockdown library were measured by quantitative high-throughput cell array phenotyping (Q-HTCP), treating with escalating doxorubicin concentrations under conditions of respiratory or glycolytic metabolism. Doxorubicin-gene interaction was quantified by departure of CPPs observed for the doxorubicin-treated mutant strain from that expected based on an interaction model. Recursive expectation-maximization clustering (REMc) and Gene Ontology (GO)-based analyses of interactions identified functional biological modules that differentially buffer or promote doxorubicin cytotoxicity with respect to Warburg metabolism. Yeast phenomic and cancer pharmacogenomics data were integrated to predict differential gene expression causally influencing doxorubicin anti-tumor efficacy. Results: Yeast compromised for genes functioning in chromatin organization, and several other cellular processes are more resistant to doxorubicin under glycolytic conditions. Thus, the Warburg transition appears to alleviate requirements for cellular functions that buffer doxorubicin cytotoxicity in a respiratory context. -

Mouse Idh3a Mutations Cause Retinal Degeneration and Reduced Mitochondrial Function Amy S
© 2018. Published by The Company of Biologists Ltd | Disease Models & Mechanisms (2018) 11, dmm036426. doi:10.1242/dmm.036426 RESEARCH ARTICLE Mouse Idh3a mutations cause retinal degeneration and reduced mitochondrial function Amy S. Findlay1, Roderick N. Carter2, Becky Starbuck3, Lisa McKie1, Klára Nováková1, Peter S. Budd1, Margaret A. Keighren1, Joseph A. Marsh1, Sally H. Cross1, Michelle M. Simon3, Paul K. Potter3, Nicholas M. Morton2 and Ian J. Jackson1,4,* ABSTRACT these diseases have a wide range of pathologies (Koopman et al., Isocitrate dehydrogenase (IDH) is an enzyme required for the 2012). Common to many of the diseases, however, are neurological production of α-ketoglutarate from isocitrate. IDH3 generates the or neuromuscular manifestations, including retinal disease. We have NADH used in the mitochondria for ATP production, and is a tetramer identified a missense mutation in the gene encoding a subunit of the made up of two α, one β and one γ subunit. Loss-of-function and tricarboxylic acid (TCA) cycle enzyme isocitrate dehydrogenase 3 missense mutations in both IDH3A and IDH3B have previously been (IDH3) as causing a retinal degeneration phenotype in mice. There implicated in families exhibiting retinal degeneration. Using mouse are three isocitrate dehydrogenase isozymes in mammals. IDH1 and models, we investigated the role of IDH3 in retinal disease and IDH2 are both homodimers that catalyse the decarboxylation of α mitochondrial function. We identified mice with late-onset retinal isocitrate to -ketoglutarate with the concomitant reduction of NADP degeneration in a screen of ageing mice carrying an ENU-induced to NADPH. IDH1 is cytoplasmic, whereas IDH2 is localised to the mutation, E229K, in Idh3a. -

Supplementary Table 1 Genes Tested in Qrt-PCR in Nfpas
Supplementary Table 1 Genes tested in qRT-PCR in NFPAs Gene Bank accession Gene Description number ABI assay ID a disintegrin-like and metalloprotease with thrombospondin type 1 motif 7 ADAMTS7 NM_014272.3 Hs00276223_m1 Rho guanine nucleotide exchange factor (GEF) 3 ARHGEF3 NM_019555.1 Hs00219609_m1 BCL2-associated X protein BAX NM_004324 House design Bcl-2 binding component 3 BBC3 NM_014417.2 Hs00248075_m1 B-cell CLL/lymphoma 2 BCL2 NM_000633 House design Bone morphogenetic protein 7 BMP7 NM_001719.1 Hs00233476_m1 CCAAT/enhancer binding protein (C/EBP), alpha CEBPA NM_004364.2 Hs00269972_s1 coxsackie virus and adenovirus receptor CXADR NM_001338.3 Hs00154661_m1 Homo sapiens Dicer1, Dcr-1 homolog (Drosophila) (DICER1) DICER1 NM_177438.1 Hs00229023_m1 Homo sapiens dystonin DST NM_015548.2 Hs00156137_m1 fms-related tyrosine kinase 3 FLT3 NM_004119.1 Hs00174690_m1 glutamate receptor, ionotropic, N-methyl D-aspartate 1 GRIN1 NM_000832.4 Hs00609557_m1 high-mobility group box 1 HMGB1 NM_002128.3 Hs01923466_g1 heterogeneous nuclear ribonucleoprotein U HNRPU NM_004501.3 Hs00244919_m1 insulin-like growth factor binding protein 5 IGFBP5 NM_000599.2 Hs00181213_m1 latent transforming growth factor beta binding protein 4 LTBP4 NM_001042544.1 Hs00186025_m1 microtubule-associated protein 1 light chain 3 beta MAP1LC3B NM_022818.3 Hs00797944_s1 matrix metallopeptidase 17 MMP17 NM_016155.4 Hs01108847_m1 myosin VA MYO5A NM_000259.1 Hs00165309_m1 Homo sapiens nuclear factor (erythroid-derived 2)-like 1 NFE2L1 NM_003204.1 Hs00231457_m1 oxoglutarate (alpha-ketoglutarate) -

Histone H3.1 (Human) Cell-Based ELISA Kit
Histone H3.1 (Human) Cell-Based ELISA Kit Catalog # : KA2761 規格 : [ 1 Kit ] List All Specification Application Image Product Histone H3.1 (Human) Cell-Based ELISA Kit is an indirect enzyme-linked Qualitative Description: immunoassay for qualitative determination of Histone H3 expression in cultured cells. Reactivity: Human, Mouse, Rat Storage Store the kit at 4°C. Instruction: Protocol: Protocol Download Suitable Attached Cell, Loosely Attached Cell, Suspension Cell Sample: Label: HRP-conjugated Detection Colorimetric Method: Regulation For research use only (RUO) Status: Datasheet: Download Applications Qualitative HIST1H3A HIST1H3D HIST1H3C HIST1H3E HIST1H3I HIST1H3G HIST1H3J HIST1H3H HIST1H3B HIST1H3F Gene Information Entrez GeneID: 8350 Protein P68431 Accession#: Gene Name: HIST1H3A Gene Alias: H3/A,H3FA Gene histone cluster 1, H3a Description: Omim ID: 602810 Gene Ontology: Hyperlink Gene Summary: Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. This structure consists of approximately 146 bp of DNA wrapped around a Page 1 of 6 2021/6/18 nucleosome, an octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a member of the histone H3 family. Transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. This gene is found in the large histone gene cluster on chromosome 6p22-p21.3. [provided by RefSeq Other H3 histone family, member A,histone 1, H3a Designations: Gene Information Entrez GeneID: 8351 Protein P68431 Accession#: Gene Name: HIST1H3D Gene Alias: H3/b,H3FB Gene histone cluster 1, H3d Description: Omim ID: 602811 Gene Ontology: Hyperlink Gene Summary: Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. -

In Silico Analysis of IDH3A Gene Revealed Novel Mutations Associated with Retinitis Pigmentosa
bioRxiv preprint doi: https://doi.org/10.1101/554196; this version posted February 18, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. In silico analysis of IDH3A gene revealed Novel mutations associated with Retinitis Pigmentosa Thwayba A. Mahmoud1*, Abdelrahman H. Abdelmoneim1, Naseem S. Murshed1, Zainab O. Mohammed2, Dina T. Ahmed1, Fatima A. Altyeb1, Nuha A. Mahmoud3, Mayada A. Mohammed1, Fatima A. Arayah1, Wafaa I. Mohammed1, Omnia S. Abayazed1, Amna S. Akasha1, Mujahed I. Mustafa1,4, Mohamed A. Hassan1 1- Department of Biotechnology, Africa city of Technology, Sudan 2- Hematology Department, Ribat University Hospital, Sudan 3- Biochemistry Department, faculty of Medicine, National University, Sudan 4- Department of Biochemistry, University of Bahri, Sudan *Corresponding Author: Thwayba A. Mahmoud, Email: [email protected] Abstract: Background: Retinitis Pigmentosa (RP) refers to a group of inherited disorders characterized by the death of photoreceptor cells leading to blindness. The aim of this study is to identify the pathogenic SNPs in the IDH3A gene and their effect on the structure and function of the protein. Method: we used different bioinformatics tools to predict the effect of each SNP on the structure and function of the protein. Result: 20 deleterious SNPs out of 178 were found to have a damaging effect on the protein structure and function. Conclusion: this is the first in silico analysis of IDH3A gene and 20 novel mutations were found using different bioinformatics tools, and they could be used as diagnostic markers for Retinitis Pigmentosa. -

A Multiprotein Occupancy Map of the Mrnp on the 3 End of Histone
Downloaded from rnajournal.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press A multiprotein occupancy map of the mRNP on the 3′ end of histone mRNAs LIONEL BROOKS III,1 SHAWN M. LYONS,2 J. MATTHEW MAHONEY,1 JOSHUA D. WELCH,3 ZHONGLE LIU,1 WILLIAM F. MARZLUFF,2 and MICHAEL L. WHITFIELD1 1Department of Genetics, Dartmouth Geisel School of Medicine, Hanover, New Hampshire 03755, USA 2Integrative Program for Biological and Genome Sciences, University of North Carolina, Chapel Hill, North Carolina 27599, USA 3Department of Computer Science, University of North Carolina, Chapel Hill, North Carolina 27599, USA ABSTRACT The animal replication-dependent (RD) histone mRNAs are coordinately regulated with chromosome replication. The RD-histone mRNAs are the only known cellular mRNAs that are not polyadenylated. Instead, the mature transcripts end in a conserved stem– loop (SL) structure. This SL structure interacts with the stem–loop binding protein (SLBP), which is involved in all aspects of RD- histone mRNA metabolism. We used several genomic methods, including high-throughput sequencing of cross-linked immunoprecipitate (HITS-CLIP) to analyze the RNA-binding landscape of SLBP. SLBP was not bound to any RNAs other than histone mRNAs. We performed bioinformatic analyses of the HITS-CLIP data that included (i) clustering genes by sequencing read coverage using CVCA, (ii) mapping the bound RNA fragment termini, and (iii) mapping cross-linking induced mutation sites (CIMS) using CLIP-PyL software. These analyses allowed us to identify specific sites of molecular contact between SLBP and its RD-histone mRNA ligands. We performed in vitro crosslinking assays to refine the CIMS mapping and found that uracils one and three in the loop of the histone mRNA SL preferentially crosslink to SLBP, whereas uracil two in the loop preferentially crosslinks to a separate component, likely the 3′hExo. -

Loss of the E3 Ubiquitin Ligase MKRN1 Represses Diet-Induced Metabolic Syndrome Through AMPK Activation
ARTICLE DOI: 10.1038/s41467-018-05721-4 OPEN Loss of the E3 ubiquitin ligase MKRN1 represses diet-induced metabolic syndrome through AMPK activation Min-Sik Lee1, Hyun-Ji Han2, Su Yeon Han2, Il Young Kim3,4, Sehyun Chae5, Choong-Sil Lee2, Sung Eun Kim2, Seul Gi Yoon4, Jun-Won Park4, Jung-Hoon Kim2, Soyeon Shin2, Manhyung Jeong2, Aram Ko2, Ho-Young Lee6, Kyoung-Jin Oh 7, Yun-Hee Lee 8, Kwang-Hee Bae7, Seung-Hoi Koo9, Jea-woo Kim10, Je Kyung Seong3,4, Daehee Hwang5 & Jaewhan Song 2 1234567890():,; AMP-activated protein kinase (AMPK) plays a key role in controlling energy metabolism in response to physiological and nutritional status. Although AMPK activation has been pro- posed as a promising molecular target for treating obesity and its related comorbidities, the use of pharmacological AMPK activators has been met with contradictory therapeutic challenges. Here we show a regulatory mechanism for AMPK through its ubiquitination and degradation by the E3 ubiquitin ligase makorin ring finger protein 1 (MKRN1). MKRN1 depletion promotes glucose consumption and suppresses lipid accumulation due to AMPK stabilisation and activation. Accordingly, MKRN1-null mice show chronic AMPK activation in both liver and adipose tissue, resulting in significant suppression of diet-induced metabolic syndrome. We demonstrate also its therapeutic effect by administering shRNA targeting MKRN1 into obese mice that reverses non-alcoholic fatty liver disease. We suggest that ubiquitin-dependent AMPK degradation represents a target therapeutic strategy for meta- bolic disorders. 1 Harvard Medical School, Boston Children’s Hospital, 3 Blackfan Circle CLS-16060.2, Boston, MA 02115, USA. 2 Department of Biochemistry, College of Life Science and Biotechnology, Yonsei University, Seoul 03722, Republic of Korea.