Gregor Mendel Freidrich Miescher Frederick Griffith
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genomics and Its Impact on Science and Society: the Human Genome Project and Beyond
DOE/SC-0083 Genomics and Its Impact on Science and Society The Human Genome Project and Beyond U.S. Department of Energy Genome Research Programs: genomics.energy.gov A Primer ells are the fundamental working units of every living system. All the instructions Cneeded to direct their activities are contained within the chemical DNA (deoxyribonucleic acid). DNA from all organisms is made up of the same chemical and physical components. The DNA sequence is the particular side-by-side arrangement of bases along the DNA strand (e.g., ATTCCGGA). This order spells out the exact instruc- tions required to create a particular organism with protein complex its own unique traits. The genome is an organism’s complete set of DNA. Genomes vary widely in size: The smallest known genome for a free-living organism (a bac- terium) contains about 600,000 DNA base pairs, while human and mouse genomes have some From Genes to Proteins 3 billion (see p. 3). Except for mature red blood cells, all human cells contain a complete genome. Although genes get a lot of attention, the proteins DNA in each human cell is packaged into 46 chro- perform most life functions and even comprise the mosomes arranged into 23 pairs. Each chromosome is majority of cellular structures. Proteins are large, complex a physically separate molecule of DNA that ranges in molecules made up of chains of small chemical com- length from about 50 million to 250 million base pairs. pounds called amino acids. Chemical properties that A few types of major chromosomal abnormalities, distinguish the 20 different amino acids cause the including missing or extra copies or gross breaks and protein chains to fold up into specific three-dimensional rejoinings (translocations), can be detected by micro- structures that define their particular functions in the cell. -

From the Human Genome Project to Genomic Medicine a Journey to Advance Human Health
From the Human Genome Project to Genomic Medicine A Journey to Advance Human Health Eric Green, M.D., Ph.D. Director, NHGRI The Origin of “Genomics”: 1987 Genomics (1987) “For the newly developing discipline of [genome] mapping/sequencing (including the analysis of the information), we have adopted the term GENOMICS… ‘The Genome Institute’ Office for Human Genome Research 1988-1989 National Center for Human Genome Research 1989-1997 National Human Genome Research Institute 1997-present NHGRI: Circa 1990-2003 Human Genome Project NHGRI Today: Characteristic Features . Relatively young (~28 years) . Relatively small (~1.7% of NIH) . Unusual historical origins (think ‘Human Genome Project’) . Emphasis on ‘Team Science’ (think managed ‘consortia’) . Rapidly disseminating footprint (think ‘genomics’) . Novel societal/bioethics research component (think ‘ELSI’) . Over-achievers for trans-NIH initiatives (think ‘Common Fund’) . Vibrant (and large) Intramural Research Program A Quarter Century of Genomics Human Genome Sequenced for First Time by the Human Genome Project Genomic Medicine An emerging medical discipline that involves using genomic information about an individual as part of their clinical care (e.g., for diagnostic or therapeutic decision- making) and the other implications of that clinical use The Path to Genomic Medicine ? Human Realization of Genome Genomic Project Medicine Nature Nature Base Pairs to Bedside 2003 Heli201x to 1Health A Quarter Century of Genomics Human Genome Sequenced for First Time by the Human Genome Project -

Nature Medicine Essay
COMMENTARY LASKER BASIC MEDICAL RESEARCH AWARD Of maize and men, or peas and people: case histories to justify plants and other model systems David Baulcombe One of the byproducts of molecular biology cork is altogether filled with air, and that air is has been support for the ‘model system’ con- perfectly enclosed in little boxes or cells distinct cept. All living organisms are based on the same from one another.”)2 (Fig. 1). Two hundred fifty genetic code, they have similar subcellular years later, Beijerinck discovered a contagium structures and they use homologous metabolic vivum fluidum in extracts of diseased tobacco pathways. So, mechanisms can be investigated plants that he later referred to as a virus3. using organisms other than those in which In contemporary science, a green alga— the knowledge will be exploited for practical Chlamydomonas reinhardtii—is a useful model benefit. Model systems are particularly use- in the analysis of kidney disease4. However, ful in the early discovery phase of a scientific in this article, I refer to the contribution of endeavor, and recent progress in biomedical plant biology to a family of mechanisms that I science has fully vindicated their use. Jacques refer to as RNA silencing. This topic has been Monod, for example, famously justified his reviewed comprehensively elsewhere5,6, so here work on a bacterial model system by stating I focus on personal experience and my view of that “what is true for Escherichia coli is also future potential from this work. true for elephants.” My fellow laureates, Victor Ambros and Gary Ruvkun, can defend the use The early history of RNA silencing in of the worm Caenorhabditis elegans as a good plants model system and so I will focus on plants. -

Next-Generation Sequencing
Tinhofer et al. Radiation Oncology (2015) 10:183 DOI 10.1186/s13014-015-0481-x REVIEW Open Access Next-generation sequencing: hype and hope for development of personalized radiation therapy? Ingeborg Tinhofer1,2*, Franziska Niehr1,2, Robert Konschak1,2, Sandra Liebs2, Matthias Munz3, Albrecht Stenzinger4, Wilko Weichert4,5, Ulrich Keilholz6 and Volker Budach1,2 Abstract The introduction of next-generation sequencing (NGS) in the field of cancer research has boosted worldwide efforts of genome-wide personalized oncology aiming at identifying predictive biomarkers and novel actionable targets. Despite considerable progress in understanding the molecular biology of distinct cancer entities by the use of this revolutionary technology and despite contemporaneous innovations in drug development, translation of NGS findings into improved concepts for cancer treatment remains a challenge. The aim of this article is to describe shortly the NGS platforms for DNA sequencing and in more detail key achievements and unresolved hurdles. A special focus will be given on potential clinical applications of this innovative technique in the field of radiation oncology. Introduction and Wong et al. [11]. We will instead focus on key achieve- Recent technological advances in DNA sequencing with ments in cancer genetics and potential clinical applications of greater speed and resolution at lower costs has provided this innovative technique in the field of radiation oncology. new insights in cancer genetics. The next-generation se- quencing (NGS) technology is tremendously facilitating the in-depth genome-wide search for genetic alterations The advantages of NGS which might significantly contribute to aggressive and/or Next-generation sequencing has rapidly been evolv- treatment-resistant phenotypes of cancers, thereby es- ing within the last decade [10]. -

Introduction and Historical Perspective
Chapter 1 Introduction and Historical Perspective “ Nothing in biology makes sense except in the light of evolution. ” modified by the developmental history of the organism, Theodosius Dobzhansky its physiology – from cellular to systems levels – and by the social and physical environment. Finally, behaviors are shaped through evolutionary forces of natural selection OVERVIEW that optimize survival and reproduction ( Figure 1.1 ). Truly, the study of behavior provides us with a window through Behavioral genetics aims to understand the genetic which we can view much of biology. mechanisms that enable the nervous system to direct Understanding behaviors requires a multidisciplinary appropriate interactions between organisms and their perspective, with regulation of gene expression at its core. social and physical environments. Early scientific The emerging field of behavioral genetics is still taking explorations of animal behavior defined the fields shape and its boundaries are still being defined. Behavioral of experimental psychology and classical ethology. genetics has evolved through the merger of experimental Behavioral genetics has emerged as an interdisciplin- psychology and classical ethology with evolutionary biol- ary science at the interface of experimental psychology, ogy and genetics, and also incorporates aspects of neuro- classical ethology, genetics, and neuroscience. This science ( Figure 1.2 ). To gain a perspective on the current chapter provides a brief overview of the emergence of definition of this field, it is helpful -

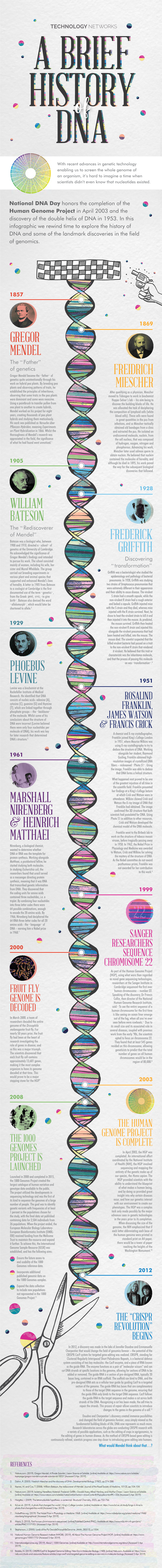
A Short History of DNA Technology 1865 - Gregor Mendel the Father of Genetics
A Short History of DNA Technology 1865 - Gregor Mendel The Father of Genetics The Augustinian monastery in old Brno, Moravia 1865 - Gregor Mendel • Law of Segregation • Law of Independent Assortment • Law of Dominance 1865 1915 - T.H. Morgan Genetics of Drosophila • Short generation time • Easy to maintain • Only 4 pairs of chromosomes 1865 1915 - T.H. Morgan •Genes located on chromosomes •Sex-linked inheritance wild type mutant •Gene linkage 0 •Recombination long aristae short aristae •Genetic mapping gray black body 48.5 body (cross-over maps) 57.5 red eyes cinnabar eyes 67.0 normal wings vestigial wings 104.5 red eyes brown eyes 1865 1928 - Frederick Griffith “Rough” colonies “Smooth” colonies Transformation of Streptococcus pneumoniae Living Living Heat killed Heat killed S cells mixed S cells R cells S cells with living R cells capsule Living S cells in blood Bacterial sample from dead mouse Strain Injection Results 1865 Beadle & Tatum - 1941 One Gene - One Enzyme Hypothesis Neurospora crassa Ascus Ascospores placed X-rays Fruiting on complete body medium All grow Minimal + amino acids No growth Minimal Minimal + vitamins in mutants Fragments placed on minimal medium Minimal plus: Mutant deficient in enzyme that synthesizes arginine Cys Glu Arg Lys His 1865 Beadle & Tatum - 1941 Gene A Gene B Gene C Minimal Medium + Citruline + Arginine + Ornithine Wild type PrecursorEnz A OrnithineEnz B CitrulineEnz C Arginine Metabolic block Class I Precursor OrnithineEnz B CitrulineEnz C Arginine Mutants Class II Mutants PrecursorEnz A Ornithine -

DNA: the Timeline and Evidence of Discovery
1/19/2017 DNA: The Timeline and Evidence of Discovery Interactive Click and Learn (Ann Brokaw Rocky River High School) Introduction For almost a century, many scientists paved the way to the ultimate discovery of DNA and its double helix structure. Without the work of these pioneering scientists, Watson and Crick may never have made their ground-breaking double helix model, published in 1953. The knowledge of how genetic material is stored and copied in this molecule gave rise to a new way of looking at and manipulating biological processes, called molecular biology. The breakthrough changed the face of biology and our lives forever. Watch The Double Helix short film (approximately 15 minutes) – hyperlinked here. 1 1/19/2017 1865 The Garden Pea 1865 The Garden Pea In 1865, Gregor Mendel established the foundation of genetics by unraveling the basic principles of heredity, though his work would not be recognized as “revolutionary” until after his death. By studying the common garden pea plant, Mendel demonstrated the inheritance of “discrete units” and introduced the idea that the inheritance of these units from generation to generation follows particular patterns. These patterns are now referred to as the “Laws of Mendelian Inheritance.” 2 1/19/2017 1869 The Isolation of “Nuclein” 1869 Isolated Nuclein Friedrich Miescher, a Swiss researcher, noticed an unknown precipitate in his work with white blood cells. Upon isolating the material, he noted that it resisted protein-digesting enzymes. Why is it important that the material was not digested by the enzymes? Further work led him to the discovery that the substance contained carbon, hydrogen, nitrogen and large amounts of phosphorus with no sulfur. -

What Is the Human Genome Project?
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Supervised Undergraduate Student Research Chancellor’s Honors Program Projects and Creative Work Spring 4-2000 What is the Human Genome Project? Lauren Leigh Taylor University of Tennessee - Knoxville Follow this and additional works at: https://trace.tennessee.edu/utk_chanhonoproj Recommended Citation Taylor, Lauren Leigh, "What is the Human Genome Project?" (2000). Chancellor’s Honors Program Projects. https://trace.tennessee.edu/utk_chanhonoproj/434 This is brought to you for free and open access by the Supervised Undergraduate Student Research and Creative Work at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Chancellor’s Honors Program Projects by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. Lauren Taylor Senior Project- (very partial) Dr.Koontz, mentor Dr. Broadhead- I should have this ready to tum in by next Tuesday or Wednesday. Thanks for your grace-- Intro As part of the University of Tennessee Honors Program, I am required to submit a senior project, consisting of research and creative analysis supervised by a faculty mentor. Although these project topics may cover any subject, most students choose a topic that falls within their undergraduate course of study. I have chosen to do this as well. As a Biology major, I have undergone ample preparation to enter a highly advanced field of modern science and medicine. One of the "hot topics" of science today is the international collaboration of scientists working to map the human genome, known as the Human Genome Project. -

Frederick Griffith and Transformation
Balderdash Example Griffith’s Transformation Experiment Ever since Edward Jenner invented the first vaccine in 1796 scientists have been working to vaccinate the world against all known diseases. Frederick Griffith wanted to save the world from pneumonia, a disease that was killing off much of Europe during the 1920’s. He didn’t build the pneumonia vaccine, but he did accidentally discover one of the most important concepts in bacterial survivability: Griffith discovered the principle of bacterial transformation. (In other words, why bacteria can fight off antibiotics) Griffith’s Transformation Experiment In 1928, Frederick Griffith was working with mice and two strains of Streptococcus pneumoniae One strain was “rough” in appearance and non-virulent, meaning that it wasn’t strong enough to hurt it’s host One strain was “smooth” in appearance and virulent. It was deadly to anyone who contracted the strain. The smooth strain looked smooth because it lacked a special protein coat that was rough in appearance and acted as a beacon summoning the mice’s immune systems. When injected with the rough (non-virulent) strain, mice lived When injected with the smooth (virulent) strain, mice died. Both as expected. Griffith’s Transformation Experiment Next, Griffith boiled the deadly, smooth strand of bacteria to kill it. He then injected mice with the deadly but boiled strand. Once again, as expected, the mice still lived. Finally, he injected the mice with BOILED smooth strands and LIVING rough strands The smooth strands are normally deadly, but Griffith had boiled them so they were not dangerous anymore. The rough strands were never deadly even when they were alive. -

Lesson Overview to Answer That Question, the First Thing You Need to 12.1 Identifying the Know Is What Genes Are Made Of
THINK ABOUT IT How do genes work? Lesson Overview To answer that question, the first thing you need to 12.1 Identifying the know is what genes are made of. Substance of Genes How would you go about figuring out what molecule or molecules go into making a gene? Griffith’s Experiments Bacterial Transformation Griffith isolated two different strains of the same bacterial The discovery of the chemical nature of the gene began in 1928 species. with British scientist Frederick Griffith, who was trying to figure Both strains grew very well in culture plates in Griffith’s lab, but out how certain types of bacteria produce pneumonia. only one of the strains caused pneumonia. The disease-causing bacteria (S strain) grew into smooth colonies on culture plates, whereas the harmless bacteria (R strain) produced colonies with rough edges. Griffith’s Experiments Griffith’s Experiments When Griffith injected mice with disease-causing bacteria, First, Griffith took a culture of the S strain, heated the cells the mice developed pneumonia and died. to kill them, and then injected the heat-killed bacteria into When he injected mice with harmless bacteria, the mice laboratory mice. stayed healthy. The mice survived, suggesting that the cause of pneumonia Perhaps the S-strain bacteria produced a toxin that made was not a toxin from these disease-causing bacteria. the mice sick? To find out, Griffith ran a series of experiments. Griffith’s Experiments Griffith’s Experiments In Griffith’s next experiment, he mixed the heat-killed, The lungs of these mice were filled with the disease-causing S-strain bacteria with live, harmless bacteria from the R bacteria. -

Long View of the Human Genome Project BOOKS & ARTS
Vol 466|19 August 2010 BOOKS & ARTS Long view of the Human Genome Project A bold attempt to tell the complicated story behind the human DNA sequence highlights that social change is needed before personalized medicine can take off, finds Jan Witkowski. Drawing the Map of Life: Inside the Human TTY E Genome Project by Victor K. McElheny Basic Books: 2010. 384 pp. $28, £16.99 S. JAFFE/AFP/G S. In 1985, Robert Sinsheimer, then chancellor of the University of California, Santa Cruz, convened a workshop to discuss sequencing the human genome. It was an audacious proposal: the longest genome that had been sequenced at the time was that of the Epstein- Barr virus, at 172,282 base pairs compared with 3 billion in human DNA. Sinsheimer’s initiative failed. Yet the idea gained momentum when, in 1988, James Watson was appointed associate director of the Office of Genome Research, part of the US National Institutes of Health (NIH). Watson declared 1990 the official start of the publicly funded NIH Human Genome Project (HGP). In 1998, Craig Venter and his company Celera Genomics, then in Rockville, Maryland, joined the race. Ten years ago in June, both projects announced a finish-line draw from President Bill Clinton’s White House. Febru- ary 2011 will mark a decade since the draft sequences were published. Genome-project pioneers: (left to right) Eric Lander, Robert Waterston, James Watson and Francis Collins. In Drawing the Map of Life, science jour- nalist and author Victor McElheny relates McElheny traces the various stages of the In 2000, HGP and Celera jointly announced the story of the HGP, from its methods to the HGP and the power struggles it engendered. -

Goings on in Mendel's Garden
40 Evolutionary Anthropology CROTCHETS & QUIDDITIES Goings on in Mendel’s Garden KENNETH WEISS The honorable monk probably didn’t cheat. But he led us astray in other ways. Gregor Mendel gave us the tools by discussed. But this may have inadver- which to do modern genetics, and we tently led us astray, in ways for which have a century of progress to show for we are paying a price today. The arti- it. We properly credit Mendel and his ficial nature of his experiments lured peas for showing us the particulate us into confusing the inheritance of nature of inheritance, but his work traits with the inheritance of genes. both enabled and disabled evolution- And this in turn has led to an unwar- ary thinking for several decades after ranted phenogenetic (see Note 1) de- its rediscovery. Since the factors he terminism that impairs our under- studied didn’t change over genera- standing of biology. tions, Mendel’s discoveries solved the problem that perplexed Darwin, that BLENDING IN blending inheritance would swamp variation and prevent evolution from Mendel wasn’t trying to explain evo- happening. Yet, for the same reason, lution. He knew of traits that varied Mendel’s work impeded evolutionary continuously in his experimental pea thought because evolution requires species, Pisum sativum, and appeared Figure 1. Mendel. change, and discrete variation was to blend in darwinian fashion from also incompatible with darwinian one generation to the next. But he gradualism. Eventually things were wanted to breed agriculturally valu- worked out, we got our unified theory able strains, so he avoided such traits used pea strains that differed only by (the neodarwinian Synthesis), and it and instead selected strains of pea the single traits he reported for each rested on Mendel’s discoveries.